- Published on
Linear Algebra
266 min read
- Authors
- Name
- Kiarash Soleimanzadeh
- https://go.kiarashs.ir/twitter
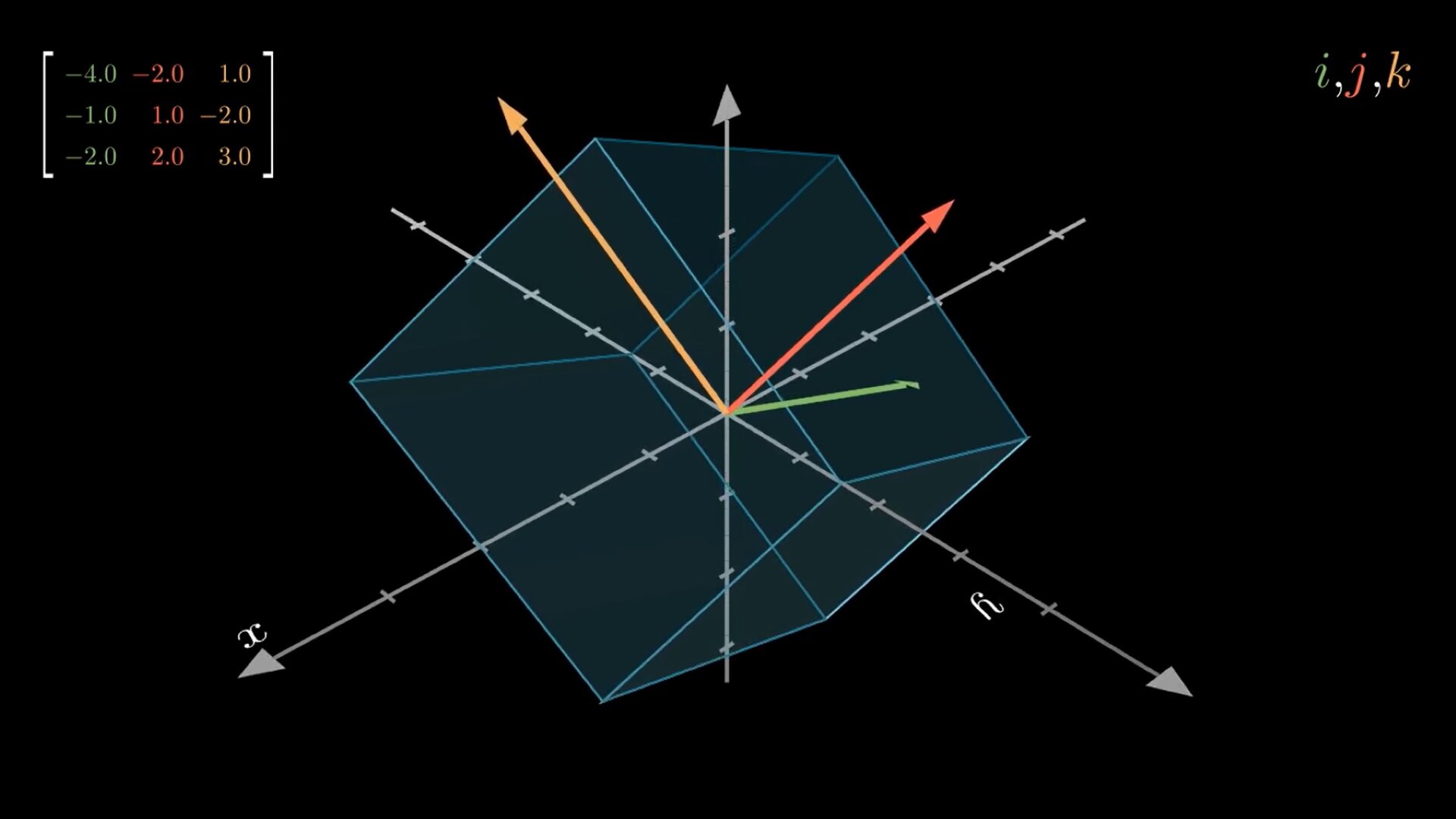
Table of Contents
- Table of contents
- Preliminary concepts
- Sets
- Belonging and inclusion
- Set specification
- Ordered pairs
- Relations
- Functions
- Vectors
- Types of vectors
- Geometric vectors
- Polynomials
- Elements of R
- Zero vector, unit vector, and sparse vector
- Vector dimensions and coordinate system
- Basic vector operations
- Vector-vector addition
- Vector-scalar multiplication
- Linear combinations of vectors
- Vector-vector multiplication: dot product
- Vector space, span, and subspace
- Vector space
- Vector span
- Vector subspaces
- Linear dependence and independence
- Vector null space
- Vector norms
- Euclidean norm
- Manhattan norm
- Max norm
- Vector inner product, length, and distance.
- Vector angles and orthogonality
- Systems of linear equations
- Matrices
- Basic Matrix operations
- Matrix-matrix addition
- Matrix-scalar multiplication
- Matrix-vector multiplication: dot product
- Matrix-matrix multiplication
- Matrix identity
- Matrix inverse
- Matrix transpose
- Hadamard product
- Special matrices
- Rectangular matrix
- Square matrix
- Diagonal matrix
- Upper triangular matrix
- Lower triangular matrix
- Symmetric matrix
- Identity matrix
- Scalar matrix
- Null or zero matrix
- Echelon matrix
- Antidiagonal matrix
- Design matrix
- Matrices as systems of linear equations
- The four fundamental matrix subsapces
- The column space
- The row space
- The null space
- The null space of the transpose
- Solving systems of linear equations with Matrices
- Gaussian Elimination
- Gauss-Jordan Elimination
- Matrix basis and rank
- Matrix norm
- Frobenius norm
- Max norm
- Spectral norm
- Linear and affine mappings
- Linear mappings
- Examples of linear mappings
- Negation matrix
- Reversal matrix
- Examples of nonlinear mappings
- Norms
- Translation
- Affine mappings
- Affine combination of vectors
- Affine span
- Affine space and subspace
- Affine mappings using the augmented matrix
- Special linear mappings
- Scaling
- Reflection
- Shear
- Rotation
- Projections
- Projections onto lines
- Projections onto general subspaces
- Projections as approximate solutions to systems of linear equations
- Matrix decompositions
- LU decomposition
- Elementary matrices
- The inverse of elementary matrices
- LU decomposition as Gaussian Elimination
- LU decomposition with pivoting
- QR decomposition
- Orthonormal basis
- Orthonormal basis transpose
- Gram-Schmidt Orthogonalization
- QR decomposition as Gram-Schmidt Orthogonalization
- Determinant
- Determinant as measures of volume
- The 2 X 2 determinant
- The N X N determinant
- Determinants as scaling factors
- The importance of determinants
- Eigenthings
- Change of basis
- Eigenvectors, Eigenvalues and Eigenspaces
- Trace and determinant with eigenvalues
- Eigendecomposition
- Eigenbasis are a good basis
- Geometric interpretation of Eigendecomposition
- The problem with Eigendecomposition
- Singular Value Decomposition
- Singular Value Decomposition Theorem
- Singular Value Decomposition computation
- Geometric interpretation of the Singular Value Decomposition
- Singular Value Decomposition vs Eigendecomposition
- Matrix Approximation
- Best rank-k approximation with SVD
- Best low-rank approximation as a minimization problem
- Some notes
- Notation
- Linearity
- Matrix properties
- Matrix calc
- Norms
- Vector norms
- Matrix norms
- Eigenstuff
- Eigenvalues intro - strang 5.1
- Strang 5.2 - diagonalization
- Strang 6.3 - singular value decomposition
- Strang 5.3 - difference eqs and power A^k
- Epilogue
Linear algebra is to machine learning as flour to bakery: every machine learning model is based in linear algebra, as every cake is based in flour. It is not the only ingredient, of course. Machine learning models need vector calculus, probability, and optimization, as cakes need sugar, eggs, and butter. Applied machine learning, like bakery, is essentially about combining these mathematical ingredients in clever ways to create useful (tasty?) models.
This document contains introductory level linear algebra notes for applied machine learning. It is meant as a reference rather than a comprehensive review. If you ever get confused by matrix multiplication, don't remember what was the norm, or the conditions for linear independence, this can serve as a quick reference. It also a good introduction for people that don't need a deep understanding of linear algebra, but still want to learn about the fundamentals to read about machine learning or to use pre-packaged machine learning solutions. Further, it is a good source for people that learned linear algebra a while ago and need a refresher.
These notes are based in a series of (mostly) freely available textbooks, video lectures, and classes I've read, watched and taken in the past. If you want to obtain a deeper understanding or to find exercises for each topic, you may want to consult those sources directly.
Free resources:
- Mathematics for Machine Learning by Deisenroth, Faisal, and Ong. 1st Ed. Book link.
- Introduction to Applied Linear Algebra by Boyd and Vandenberghe. 1sr Ed. Book link
- Linear Algebra Ch. in Deep Learning by Goodfellow, Bengio, and Courville. 1st Ed. Chapter link.
- Linear Algebra Ch. in Dive into Deep Learning by Zhang, Lipton, Li, And Smola. Chapter link.
- Prof. Pavel Grinfeld's Linear Algebra Lectures at Lemma. Videos link.
- Prof. Gilbert Strang's Linear Algebra Lectures at MIT. Videos link.
- Salman Khan's Linear Algebra Lectures at Khan Academy. Videos link.
- 3blue1brown's Linear Algebra Series at YouTube. Videos link.
Not-free resources:
- Introduction to Linear Algebra by Gilbert Strang. 5th Ed. Book link.
- No Bullshit Guide to Linear Algebra by Ivan Savov. 2nd Ed. Book Link.
I've consulted all these resources at one point or another. Pavel Grinfeld's lectures are my absolute favorites. Salman Khan's lectures are really good for absolute beginners (they are long though). The famous 3blue1brown series in linear algebra is delightful to watch and to get a solid high-level view of linear algebra.
If you have to pic one book, I'd pic Boyd's and Vandenberghe's Intro to applied linear algebra, as it is the most beginner friendly book on linear algebra I've encounter. Every aspect of the notation is clearly explained and pretty much all the key content for applied machine learning is covered. The Linear Algebra Chapter in Goodfellow et al is a nice and concise introduction, but it may require some previous exposure to linear algebra concepts. Deisenroth et all book is probably the best and most comprehensive source for linear algebra for machine learning I've found, although it assumes that you are good at reading math (and at math more generally). Savov's book it's also great for beginners but requires time to digest. Professor Strang lectures are great too but I won't recommend it for absolute beginners.
I'll do my best to keep notation consistent. Nevertheless, learning to adjust to changing or inconsistent notation is a useful skill, since most authors will use their own preferred notation, and everyone seems to think that its/his/her own notation is better.
To make everything more dynamic and practical, I'll introduce bits of Python code to exemplify each mathematical operation (when possible) with NumPy, which is the facto standard package for scientific computing in Python.
Finally, keep in mind this is created by a non-mathematician for (mostly) non-mathematicians. I wrote this as if I were talking to myself or a dear friend, which explains why my writing is sometimes conversational and informal.
If you find any mistake in notes feel free to reach me, so I can correct the issue.
Table of contents
Note: underlined sections are the newest sections and/or corrected ones.
- Types of vectors
- Zero vector, unit vector, and sparse vector
- Vector dimensions and coordinate system
- Basic vector operations
- Vector space, span, and subspace
- Linear dependence and independence
- Vector null space
- Vector norms
- Vector inner product, length, and distance
- Vector angles and orthogonality
- Systems of linear equations
- Basic matrix operations
- Special matrices
- Matrices as systems of linear equations
- The four fundamental matrix subsapces
- Solving systems of linear equations with matrices
- Matrix basis and rank
- Matrix norm
- Linear mappings
- Examples of linear mappings
- Examples of nonlinear mappings
- Affine mappings
- Special linear mappings
- Projections
- LU decomposition
- QR decomposition
- Determinant
- Eigenthings
- Singular Value Decomposition:
- Matrix Approximation:
- Some notes
Preliminary concepts
While writing about linear mappings, I realized the importance of having a basic understanding of a few concepts before approaching the study of linear algebra. If you are like me, you may not have formal mathematical training beyond high school. If so, I encourage you to read this section and spent some time wrapping your head around these concepts before going over the linear algebra content (otherwise, you might prefer to skip this part). I believe that reviewing these concepts is of great help to understand the notation, which in my experience is one of the main barriers to understand mathematics for nonmathematicians: we are nonnative speakers, so we are continuously building up our vocabulary. I'll keep this section very short, as is not the focus of this mini-course.
For this section, my notes are based on readings of:
- Geometric transformations (Vol. 1) (1966) by Modenov & Parkhomenko
- Naive Set Theory (1960) by P.R. Halmos
- Abstract Algebra: Theory and Applications (2016) by Judson & Beeer. Book link
Sets
Sets are one of the most fundamental concepts in mathematics. They are so fundamentals that they are not defined in terms of anything else. On the contrary, other branches of mathematics are defined in terms of sets, including linear algebra. Put simply, sets are well-defined collections of objects. Such objects are called elements or members of the set. The crew of a ship, a caravan of camels, and the LA Lakers roster, are all examples of sets. The captain of the ship, the first camel in the caravan, and LeBron James are all examples of "members" or "elements" of their corresponding sets. We denote a set with an upper case italic letter as . In the context of linear algebra, we say that a line is a set of points, and the set of all lines in the plane is a set of sets. Similarly, we can say that vectors are sets of points, and matrices sets of vectors.
Belonging and inclusion
We build sets using the notion of belonging. We denote that belongs (or is an element or member of) to with the Greek letter epsilon as:
Another important idea is inclusion, which allow us to build subsets. Consider sets and . When every element of is an element of , we say that is a subset of , or that includes . The notation is:
or
Belonging and inclusion are derived from axion of extension: two sets are equal if and only if they have the same elements. This axiom may sound trivially obvious but is necessary to make belonging and inclusion rigorous.
Set specification
In general, anything we assert about the elements of a set results in generating a subset. In other words, asserting things about sets is a way to manufacture subsets. Take as an example the set of all dogs, that I'll denote as . I can assert now " is black". Such an assertion is true for some members of the set of all dogs and false for others. Hence, such a sentence, evaluated for all member of , generates a subset: the set of all black dogs. This is denoted as:
or
The colon () or vertical bar () read as "such that". Therefore, we can read the above expression as: all elements of in such that is black. And that's how we obtain the set from .
Set generation, as defined before, depends on the axiom of specification: to every set and to every condition there corresponds a set whose elements are exactly those elements for which holds.
A condition is any sentence or assertion about elements of . Valid sentences are either of belonging or equality. When we combine belonging and equality assertions with logic operators (not, if, and or, etc), we can build any legal set.
Ordered pairs
Pairs of sets come in two flavors: unordered and ordered. We care about pairs of sets as we need them to define a notion of relations and functions (from here I'll denote sets with lower-case for convenience, but keep in mind we're still talking about sets).
Consider a pair of sets and . An unordered pair is a set whose elements are , and . Therefore, presentation order does not matter, the set is the same.
In machine learning, we usually do care about presentation order. For this, we need to define an ordered pair (I'll introduce this at an intuitive level, to avoid to introduce too many new concepts). An ordered pair is denoted as , with as the first coordinate and as the second coordinate. A valid ordered pair has the property that .
Relations
From ordered pairs, we can derive the idea of relations among sets or between elements and sets. Relations can be binary, ternary, quaternary, or N-ary. Here we are just concerned with binary relationships. In set theory, relations are defined as sets of ordered pairs, and denoted as . Hence, we can express the relation between and as:
Further, for any , there exist and such that .
From the definition of , we can obtain the notions of domain and range. The domain is a set defined as:
This reads as: the values of such that for at least one element of , has a relation with .
The range is a set defined as:
This reads: the set formed by the values of such that at least one element of , has a relation with .
Functions
Consider a pair of sets and . We say that a function from to is relation such that:
- and
- such that for each there is a unique element of with
More informally, we say that a function "transform" or "maps" or "sends" onto , and for each "argument" there is a unique value that "assummes" or "takes".
We typically denote a relation or function or transformation or mapping from X onto Y as:
or
The simples way to see the effect of this definition of a function is with a chart. In Fig. 1, the left-pane shows a valid function, i.e., each value maps uniquely onto one value of . The right-pane is not a function, since each value maps onto multiple values of .
For , the domain of equals to , but the range does not necessarily equals to . Just recall that the range includes only the elements for which has a relation with .
The ultimate goal of machine learning is learning functions from data, i.e., transformations or mappings from the domain onto the range of a function. This may sound simplistic, but it's true. The domain is usually a vector (or set) of variables or features mapping onto a vector of target values. Finally, I want to emphasize that in machine learning the words transformation and mapping are used interchangeably, but both just mean function.
This is all I'll cover about sets and functions. My goals were just to introduce: (1) the concept of a set, (2) basic set notation, (3) how sets are generated, (4) how sets allow the definition of functions, (5) the concept of a function. Set theory is a monumental field, but there is no need to learn everything about sets to understand linear algebra. Halmo's Naive set theory (not free, but you can find a copy for ~\10 US) is a fantastic book for people that just need to understand the most fundamental ideas in a relatively informal manner.
# Libraries for this section
import numpy as np
import pandas as pd
import altair as alt
alt.themes.enable('dark')
# ThemeRegistry.enable('dark')
Vectors
Linear algebra is the study of vectors. At the most general level, vectors are ordered finite lists of numbers. Vectors are the most fundamental mathematical object in machine learning. We use them to represent attributes of entities: age, sex, test scores, etc. We represent vectors by a bold lower-case letter like or as a lower-case letter with an arrow on top like .
Vectors are a type of mathematical object that can be added together and/or multiplied by a number to obtain another object of the same kind. For instance, if we have a vector and a second vector , we can add them together and obtain a third vector . We can also multiply to obtain , again, a vector. This is what we mean by the same kind: the returning object is still a vector.
Types of vectors
Vectors come in three flavors: (1) geometric vectors, (2) polynomials, (3) and elements of space. We will defined each one next.
Geometric vectors
Geometric vectors are oriented segments. Therse are the kind of vectors you probably learned about in high-school physics and geometry. Many linear algebra concepts come from the geometric point of view of vectors: space, plane, distance, etc.
Polynomials
A polynomial is an expression like . This is, a expression adding multiple "terms" (nomials). Polynomials are vectors because they meet the definition of a vector: they can be added together to get another polynomial, and they can be multiplied together to get another polynomial.
Elements of R
Elements of are sets of real numbers. This type of representation is arguably the most important for applied machine learning. It is how data is commonly represented in computers to build machine learning models. For instance, a vector in takes the shape of:
Indicating that it contains three dimensions.
In NumPy vectors are represented as n-dimensional arrays. To create a vector in :
x = np.array([[1],
[2],
[3]])
We can inspect the vector shape by:
x.shape # (3 dimensions, 1 element on each)
# (3, 1)
print(f'A 3-dimensional vector:\n{x}')
"""
A 3-dimensional vector:
[[1]
[2]
[3]]
"""
Zero vector, unit vector, and sparse vector
There are a couple of "special" vectors worth to remember as they will be mentioned frequently on applied linear algebra: (1) zero vector, (2) unit vector, (3) sparse vectors
Zero vectors, are vectors composed of zeros, and zeros only. It is common to see this vector denoted as simply , regardless of the dimensionality. Hence, you may see a 3-dimensional or 10-dimensional with all entries equal to 0, refered as "the 0" vector. For instance:
Unit vectors, are vectors composed of a single element equal to one, and the rest to zero. Unit vectors are important to understand applications like norms. For instance, , , and are unit vectors:
Sparse vectors, are vectors with most of its elements equal to zero. We denote the number of nonzero elements of a vector as . The sparser possible vector is the zero vector. Sparse vectors are common in machine learning applications and often require some type of method to deal with them effectively.
Vector dimensions and coordinate system
Vectors can have any number of dimensions. The most common are the 2-dimensional cartesian plane, and the 3-dimensional space. Vectors in 2 and 3 dimensions are used often for pedgagogical purposes since we can visualize them as geometric vectors. Nevetheless, most problems in machine learning entail more dimensions, sometiome hundreds or thousands of dimensions. The notation for a vector of arbitrary dimensions, is:
Vectors dimensions map into coordinate systems or perpendicular axes. Coordinate systems have an origin at , hence, when we define a vector:
we are saying: starting from the origin, move 3 units in the 1st perpendicular axis, 2 units in the 2nd perpendicular axis, and 1 unit in the 3rd perpendicular axis. We will see later that when we have a set of perpendicular axes we obtain the basis of a vector space.
Basic vector operations
Vector-vector addition
We used vector-vector addition to define vectors without defining vector-vector addition. Vector-vector addition is an element-wise operation, only defined for vectors of the same size (i.e., number of elements). Consider two vectors of the same size, then:
For instance:
Vector addition has a series of fundamental properties worth mentioning:
- Commutativity:
- Associativity:
- Adding the zero vector has no effect:
- Substracting a vector from itself returns the zero vector:
In NumPy, we add two vectors of the same with the + operator or the add method:
x = y = np.array([[1],
[2],
[3]])
x + y
"""
array([[2],
[4],
[6]])
"""
np.add(x,y)
"""
array([[2],
[4],
[6]])
"""
Vector-scalar multiplication
Vector-scalar multiplication is an element-wise operation. It's defined as:
Consider and :
Vector-scalar multiplication satisfies a series of important properties:
- Associativity:
- Left-distributive property:
- Right-distributive property:
- Right-distributive property for vector addition:
In NumPy, we compute scalar-vector multiplication with the * operator:
alpha = 2
x = np.array([[1],
[2],
[3]])
alpha * x
"""
array([[2],
[4],
[6]])
"""
Linear combinations of vectors
There are only two legal operations with vectors in linear algebra: addition and multiplication by numbers. When we combine those, we get a linear combination.
Consider , , , and .
We obtain:
Another way to express linear combinations you'll see often is with summation notation. Consider a set of vectors and scalars , then:
Note that means "is defined as".
Linear combinations are the most fundamental operation in linear algebra. Everything in linear algebra results from linear combinations. For instance, linear regression is a linear combination of vectors. Fig. 2 shows an example of how adding two geometrical vectors looks like for intuition.
In NumPy, we do linear combinations as:
a, b = 2, 3
x , y = np.array([[2],[3]]), np.array([[4], [5]])
a*x + b*y
"""
array([[16],
[21]])
"""
Vector-vector multiplication: dot product
We covered vector addition and multiplication by scalars. Now I will define vector-vector multiplication, commonly known as a dot product or inner product. The dot product of and is defined as:
Where the superscript denotes the transpose of the vector. Transposing a vector just means to "flip" the column vector to a row vector counterclockwise. For instance:
Dot products are so important in machine learning, that after a while they become second nature for practitioners.
To multiply two vectors with dimensions (rows=2, cols=1) in Numpy, we need to transpose the first vector at using the @ operator:
x, y = np.array([[-2],[2]]), np.array([[4],[-3]])
x.T @ y
# array([[-14]])
Vector space, span, and subspace
Vector space
In its more general form, a vector space, also known as linear space, is a collection of objects that follow the rules defined for vectors in . We mentioned those rules when we defined vectors: they can be added together and multiplied by scalars, and return vectors of the same type. More colloquially, a vector space is the set of proper vectors and all possible linear combinatios of the vector set. In addition, vector addition and multiplication must follow these eight rules:
- commutativity:
- associativity:
- unique zero vector such that:
- there is a unique vector such that
- identity element of scalar multiplication:
- distributivity of scalar multiplication w.r.t vector addition:
In my experience remembering these properties is not really important, but it's good to know that such rules exist.
Vector span
Consider the vectors and and the scalars and . If we take all possible linear combinations of we would obtain the span of such vectors. This is easier to grasp when you think about geometric vectors. If our vectors and point into different directions in the 2-dimensional space, we get that the is equal to the entire 2-dimensional plane, as shown in the middle-pane in Fig. 5. Just imagine having an unlimited number of two types of sticks: one pointing vertically, and one pointing horizontally. Now, you can reach any point in the 2-dimensional space by simply combining the necessary number of vertical and horizontal sticks (including taking fractions of sticks).
What would happen if the vectors point in the same direction? Now, if you combine them, you just can span a line, as shown in the left-pane in Fig. 5. If you have ever heard of the term "multicollinearity", it's closely related to this issue: when two variables are "colinear" they are pointing in the same direction, hence they provide redundant information, so can drop one without information loss.
With three vectors pointing into different directions, we can span the entire 3-dimensional space or a hyper-plane, as in the right-pane of Fig. 5. Note that the sphere is just meant as a 3-D reference, not as a limit.
Four vectors pointing into different directions will span the 4-dimensional space, and so on. From here our geometrical intuition can't help us. This is an example of how linear algebra can describe the behavior of vectors beyond our basics intuitions.
Vector subspaces
A vector subspace (or linear subspace) is a vector space that lies within a larger vector space. These are also known as linear subspaces. Consider a subspace . For a vector to be a valid subspace it has to meet three conditions:
- Contains the zero vector,
- Closure under multiplication,
- Closure under addition,
Intuitively, you can think in closure as being unable to "jump out" from space into another. A pair of vectors laying flat in the 2-dimensional space, can't, by either addition or multiplication, "jump out" into the 3-dimensional space.
Consider the following questions: Is a valid subspace of ? Let's evaluate on the three conditions:
Contains the zero vector: it does. Remember that the span of a vector are all linear combinations of such a vector. Therefore, we can simply multiply by to get :
Closure under multiplication implies that if take any vector belonging to and multiply by any real scalar , the resulting vector stays within the span of . Algebraically is easy to see that we can multiply by any scalar , and the resulting vector remains in the 2-dimensional plane (i.e., the span of ).
Closure under addition implies that if we add together any vectors belonging to , the resulting vector remains within the span of . Again, algebraically is clear that if we add + , the resulting vector will remain in . There is no way to get to or or any space outside the two-dimensional plane by adding multiple times.
Linear dependence and independence
The left-pane shows a triplet of linearly dependent vectors, whereas the right-pane shows a triplet of linearly independent vectors.
A set of vectors is linearly dependent if at least one vector can be obtained as a linear combination of other vectors in the set. As you can see in the left pane, we can combine vectors and to obtain .
There is more rigurous (but slightly harder to grasp) definition of linear dependence. Consider a set of vectors and scalars . If there is a way to get with at least one , we have linearly dependent vectors. In other words, if we can get the zero vector as a linear combination of the vectors in the set, with weights that are not all zero, we have a linearly dependent set.
A set of vectors is linearly independent if none vector can be obtained as a linear combination of other vectors in the set. As you can see in the right pane, there is no way for us to combine vectors and to obtain . Again, consider a set of vectors and scalars . If the only way to get requires all , the we have linearly independent vectors. In words, the only way to get the zero vectors in by multoplying each vector in the set by .
The importance of the concepts of linear dependence and independence will become clearer in more advanced topics. For now, the important points to remember are: linearly dependent vectors contain redundant information, whereas linearly independent vectors do not.
Vector null space
Now that we know what subspaces and linear dependent vectors are, we can introduce the idea of the null space. Intuitively, the null space of a set of vectors are all linear combinations that "map" into the zero vector. Consider a set of geometric vectors , , , and as in Fig. 8. By inspection, we can see that vectors and are parallel to each other, hence, independent. On the contrary, vectors and can be obtained as linear combinations of and , therefore, dependent.
As result, with this four vectors, we can form the following two combinations that will "map" into the origin of the coordinate system, this is, the zero vector :
We will see how this idea of the null space extends naturally in the context of matrices later.
Vector norms
Measuring vectors is another important operation in machine learning applications. Intuitively, we can think about the norm or the length of a vector as the distance between its "origin" and its "end".
Norms "map" vectors to non-negative values. In this sense are functions that assign length to a vector . To be valid, a norm has to satisfy these properties (keep in mind these properties are a bit abstruse to understand):
- Absolutely homogeneous: . In words: for all real-valued scalars, the norm scales proportionally with the value of the scalar.
- Triangle inequality: . In words: in geometric terms, for any triangle the sum of any two sides must be greater or equal to the lenght of the third side. This is easy to see experimentally: grab a piece of rope, form triangles of different sizes, measure all the sides, and test this property.
- Positive definite: and . In words: the length of any has to be a positive value (i.e., a vector can't have negative length), and a length of occurs only of
Grasping the meaning of these three properties may be difficult at this point, but they probably become clearer as you improve your understanding of linear algebra.
Euclidean norm
The Euclidean norm is one of the most popular norms in machine learning. It is so widely used that sometimes is refered simply as "the norm" of a vector. Is defined as:
Hence, in two dimensions the norm is:
Which is equivalent to the formula for the hypotenuse a triangle with sides and .
The same pattern follows for higher dimensions of
In NumPy, we can compute the norm as:
x = np.array([[3],[4]])
np.linalg.norm(x, 2)
# 5.0
If you remember the first "Pythagorean triple", you can confirm that the norm is correct.
Manhattan norm
The Manhattan or norm gets its name in analogy to measuring distances while moving in Manhattan, NYC. Since Manhattan has a grid-shape, the distance between any two points is measured by moving in vertical and horizontals lines (instead of diagonals as in the Euclidean norm). It is defined as:
Where is the absolute value. The norm is preferred when discriminating between elements that are exactly zero and elements that are small but not zero.
In NumPy we compute the norm as
x = np.array([[3],[-4]])
np.linalg.norm(x, 1)
# 7.0
Is easy to confirm that the sum of the absolute values of and is .
Max norm
The max norm or infinity norm is simply the absolute value of the largest element in the vector. It is defined as:
Where is the absolute value. For instance, for a vector with elements , the
In NumPy we compute the norm as:
x = np.array([[3],[-4]])
np.linalg.norm(x, np.inf)
# 4.0
Vector inner product, length, and distance.
For practical purposes, inner product and length are used as equivalent to dot product and norm, although technically are not the same.
Inner products are a more general concept that dot products, with a series of additional properties (see here). In other words, every dot product is an inner product, but not every inner product is a dot product. The notation for the inner product is usually a pair of angle brackets as as. For instance, the scalar inner product is defined as:
In the inner product is a dot product defined as:
Length is a concept from geometry. We say that geometric vectors have length and that vectors in have norm. In practice, many machine learning textbooks use these concepts interchangeably. I've found authors saying things like "we use the norm to compute the length of a vector". For instance, we can compute the length of a directed segment (i.e., geometrical vector) by taking the square root of the inner product with itself as:
Distance is a relational concept. It refers to the length (or norm) of the difference between two vectors. Hence, we use norms and lengths to measure the distance between vectors. Consider the vectors and , we define the distance as:
When the inner product is the dot product, the distance equals to the Euclidean distance.
In machine learning, unless made explicit, we can safely assume that an inner product refers to the dot product. We already reviewed how to compute the dot product in NumPy:
x, y = np.array([[-2],[2]]), np.array([[4],[-3]])
x.T @ y
# array([[-14]])
As with the inner product, usually, we can safely assume that distance stands for the Euclidean distance or norm unless otherwise noted. To compute the distance between a pair of vectors:
distance = np.linalg.norm(x-y, 2)
print(f'L_2 distance : {distance}')
# L_2 distance : 7.810249675906656
Vector angles and orthogonality
The concepts of angle and orthogonality are also related to geometrical vectors. We saw that inner products allow for the definition of length and distance. In the same manner, inner products are used to define angles and orthogonality.
In machine learning, the angle between a pair of vectors is used as a measure of vector similarity. To understand angles let's first look at the Cauchy–Schwarz inequality. Consider a pair of non-zero vectors and . The Cauchy–Schwarz inequality states that:
In words: the absolute value of the inner product of a pair of vectors is less than or equal to the products of their length. The only case where both sides of the expression are equal is when vectors are colinear, for instance, when is a scaled version of . In the 2-dimensional case, such vectors would lie along the same line.
The definition of the angle between vectors can be thought as a generalization of the law of cosines in trigonometry, which defines for a triangle with sides , , and , and an angle are related as:
We can replace this expression with vectors lengths as:
With a bit of algebraic manipulation, we can clear the previous equation to:
And there we have a definition for (cos) angle . Further, from the Cauchy–Schwarz inequality we know that must be:
This is a necessary conclusion (range between ) since the numerator in the equation always is going to be smaller or equal to the denominator.
In NumPy, we can compute the between a pair of vectors as:
x, y = np.array([[1], [2]]), np.array([[5], [7]])
# here we translate the cos(theta) definition
cos_theta = (x.T @ y) / (np.linalg.norm(x,2) * np.linalg.norm(y,2))
print(f'cos of the angle = {np.round(cos_theta, 3)}')
# cos of the angle = [[0.988]]
We get that . Finally, to know the exact value of we need to take the trigonometric inverse of the cosine function as:
cos_inverse = np.arccos(cos_theta)
print(f'angle in radiants = {np.round(cos_inverse, 3)}')
# angle in radiants = [[0.157]]
We obtain . To fo from radiants to degrees we can use the following formula:
degrees = cos_inverse * ((180)/np.pi)
print(f'angle in degrees = {np.round(degrees, 3)}')
# angle in degrees = [[8.973]]
We obtain
Orthogonality is often used interchangeably with "independence" although they are mathematically different concepts. Orthogonality can be seen as a generalization of perpendicularity to vectors in any number of dimensions.
We say that a pair of vectors and are orthogonal if their inner product is zero, . The notation for a pair of orthogonal vectors is . In the 2-dimensional plane, this equals to a pair of vectors forming a angle.
Here is an example of orthogonal vectors
x = np.array([[2], [0]])
y = np.array([[0], [2]])
cos_theta = (x.T @ y) / (np.linalg.norm(x,2) * np.linalg.norm(y,2))
print(f'cos of the angle = {np.round(cos_theta, 3)}')
# cos of the angle = [[0.]]
We see that this vectors are orthogonal as . This is equal to radiants and
cos_inverse = np.arccos(cos_theta)
degrees = cos_inverse * ((180)/np.pi)
print(f'angle in radiants = {np.round(cos_inverse, 3)}\nangle in degrees ={np.round(degrees, 3)} ')
"""
angle in radiants = [[1.571]]
angle in degrees =[[90.]]
"""
Systems of linear equations
The purpose of linear algebra as a tool is to solve systems of linear equations. Informally, this means to figure out the right combination of linear segments to obtain an outcome. Even more informally, think about making pancakes: In what proportion () we have to mix ingredients to make pancakes? You can express this as a linear equation:
The above expression describe a linear equation. A system of linear equations involve multiple equations that have to be solved simultaneously. Consider:
Now we have a system with two unknowns, and . We'll see general methods to solve systems of linear equations later. For now, I'll give you the answer: and . Geometrically, we can see that both equations produce a straight line in the 2-dimensional plane. The point on where both lines encounter is the solution to the linear system.
df = pd.DataFrame({"x1": [0, 2], "y1":[8, 3], "x2": [0.5, 2], "y2": [0, 3]})
equation1 = alt.Chart(df).mark_line().encode(x="x1", y="y1")
equation2 = alt.Chart(df).mark_line(color="red").encode(x="x2", y="y2")
equation1 + equation2
Matrices
# Libraries for this section
import numpy as np
import pandas as pd
import altair as alt
Matrices are as fundamental as vectors in machine learning. With vectors, we can represent single variables as sets of numbers or instances. With matrices, we can represent sets of variables. In this sense, a matrix is simply an ordered collection of vectors. Conventionally, column vectors, but it's always wise to pay attention to the authors' notation when reading matrices. Since computer screens operate in two dimensions, matrices are the way in which we interact with data in practice.
More formally, we represent a matrix with a italicized upper-case letter like . In two dimensions, we say the matrix has rows and columns. Each entry of is defined as , and . A matrix is defines as:
In Numpy, we construct matrices with the array method:
A = np.array([[0,2], # 1st row
[1,4]]) # 2nd row
print(f'a 2x2 Matrix:\n{A}')
"""
a 2x2 Matrix:
[[0 2]
[1 4]]
"""
Basic Matrix operations
Matrix-matrix addition
We add matrices in a element-wise fashion. The sum of and is defined as:
For instance:
In Numpy, we add matrices with the + operator or add method:
A = np.array([[0,2],
[1,4]])
B = np.array([[3,1],
[-3,2]])
A + B
"""
array([[ 3, 3],
[-2, 6]])
"""
np.add(A, B)
"""
array([[ 3, 3],
[-2, 6]])
"""
Matrix-scalar multiplication
Matrix-scalar multiplication is an element-wise operation. Each element of the matrix is multiplied by the scalar . Is defined as:
Consider and , then:
In NumPy, we compute matrix-scalar multiplication with the * operator or multiply method:
alpha = 2
A = np.array([[1,2],
[3,4]])
alpha * A
"""
array([[2, 4],
[6, 8]])
"""
np.multiply(alpha, A)
"""
array([[2, 4],
[6, 8]])
"""
Matrix-vector multiplication: dot product
Matrix-vector multiplication equals to taking the dot product of each column of a with each element resulting in a vector . Is defined as:
For instance:
In numpy, we compute the matrix-vector product with the @ operator or dot method:
A = np.array([[0,2],
[1,4]])
x = np.array([[1],
[2]])
A @ x
"""
array([[4],
[9]])
"""
np.dot(A, x)
"""
array([[4],
[9]])
"""
Matrix-matrix multiplication
Matrix-matrix multiplication is a dot produt as well. To work, the number of columns in the first matrix has to be equal to the number of rows in the second matrix . Hence, times to be valid. One way to see matrix-matrix multiplication is by taking a series of dot products: the 1st column of times the 1st row of , the 2nd column of times the 2nd row of , until the column of times the row of .
We define :
A compact way to define the matrix-matrix product is:
For instance
Matrix-matrix multiplication has a series of important properties:
- Associativity:
- Associativity with scalar multiplication:
- Distributivity with addition:
- Transpose of product:
It's also important to remember that matrix-matrix multiplication orders matter, this is, it is not commutative. Hence, in general, .
In NumPy, we obtan the matrix-matrix product with the @ operator or dot method:
A = np.array([[0,2],
[1,4]])
B = np.array([[1,3],
[2,1]])
A @ B
"""
array([[4, 2],
[9, 7]])
"""
np.dot(A, B)
"""
array([[4, 2],
[9, 7]])
"""
Matrix identity
An identity matrix is a square matrix with ones on the diagonal from the upper left to the bottom right, and zeros everywhere else. We denote the identity matrix as . We define as:
For example:
You can think in the inverse as playing the same role than in operations with real numbers. The inverse matrix does not look very interesting in itself, but it plays an important role in some proofs and for the inverse matrix (which can be used to solve system of linear equations).
Matrix inverse
In the context of real numbers, the multiplicative inverse (or reciprocal) of a number , is the number that when multiplied by yields . We denote this by or . Take the number . Its multiplicative inverse equals to .
If you recall the matrix identity section, we said that the identity plays a similar role than the number one but for matrices. Again, by analogy, we can see the inverse of a matrix as playing the same role than the multiplicative inverse for numbers but for matrices. Hence, the inverse matrix is a matrix than when multiplies another matrix from either the right or the left side, returns the identity matrix.
More formally, consider the square matrix . We define as matrix with the property:
The main reason we care about the inverse, is because it allows to solve systems of linear equations in certain situations. Consider a system of linear equations as:
Assuming has an inverse, we can multiply by the inverse on both sides:
And get:
Since the does not affect at all, our final expression becomes:
This means that we just need to know the inverse of , multiply by the target vector , and we obtain the solution for our system. I mentioned that this works only in certain situations. By this I meant: if and only if happens to have an inverse. Not all matrices have an inverse. When exist, we say is nonsingular or invertible, otherwise, we say it is noninvertible or singular.
The lingering question is how to find the inverse of a matrix. We can do it by reducing to its reduced row echelon form by using Gauss-Jordan Elimination. If has an inverse, we will obtain the identity matrix as the row echelon form of . I haven't introduced either just yet. You can jump to the Solving systems of linear equations with matrices if you are eager to learn about it now. For now, we relie on NumPy.
In NumPy, we can compute the inverse of a matrix with the .linalg.inv method:
A = np.array([[1, 2, 1],
[4, 4, 5],
[6, 7, 7]])
A_i = np.linalg.inv(A)
print(f'A inverse:\n{A_i}')
"""
A inverse:
[[-7. -7. 6.]
[ 2. 1. -1.]
[ 4. 5. -4.]]
"""
We can check the is correct by multiplying. If so, we should obtain the identity
I = np.round(A_i @ A)
print(f'A_i times A resulsts in I_3:\n{I}')
"""
A_i times A resulsts in I_3:
[[ 1. 0. 0.]
[ 0. 1. -0.]
[ 0. -0. 1.]]
"""
Matrix transpose
Consider a matrix . The transpose of is denoted as . We obtain as:
In other words, we get the by switching the columns by the rows of . For instance:
In NumPy, we obtain the transpose with the T method:
A = np.array([[1, 2],
[3, 4],
[5, 6]])
A.T
"""
array([[1, 3, 5],
[2, 4, 6]])
"""
Hadamard product
It is tempting to think in matrix-matrix multiplication as an element-wise operation, as multiplying each overlapping element of and . It is not. Such operation is called Hadamard product. I'm introducing this to avoid confusion. The Hadamard product is defined as
For instance:
In numpy, we compute the Hadamard product with the * operator or multiply method:
A = np.array([[0,2],
[1,4]])
B = np.array([[1,3],
[2,1]])
A * B
"""
array([[0, 6],
[2, 4]])
"""
np.multiply(A, B)
"""
array([[0, 6],
[2, 4]])
"""
Special matrices
There are several matrices with special names that are commonly found in machine learning theory and applications. Knowing these matrices beforehand can improve your linear algebra fluency, so we will briefly review a selection of 12 common matrices. For an extended list of special matrices see here and here.
Rectangular matrix
Matrices are said to be rectangular when the number of rows is to the number of columns, i.e., with . For instance:
Square matrix
Matrices are said to be square when the number of rows the number of columns, i.e., . For instance:
Diagonal matrix
Square matrices are said to be diagonal when each of its non-diagonal elements is zero, i.e., For , we have . For instance:
Upper triangular matrix
Square matrices are said to be upper triangular when the elements below the main diagonal are zero, i.e., For , we have . For instance:
Lower triangular matrix
Square matrices are said to be lower triangular when the elements above the main diagonal are zero, i.e., For , we have . For instance:
Symmetric matrix
Square matrices are said to be symmetric its equal to its transpose, i.e., . For instance:
Identity matrix
A diagonal matrix is said to be the identity when the elements along its main diagonal are equal to one. For instance:
Scalar matrix
Diagonal matrices are said to be scalar when all the elements along its main diaonal are equal, i.e., . For instance:
Null or zero matrix
Matrices are said to be null or zero matrices when all its elements equal to zero, wich is denoted as . For instance:
Echelon matrix
Matrices are said to be on echelon form when it has undergone the process of Gaussian elimination. More specifically:
- Zero rows are at the bottom of the matrix
- The leading entry (pivot) of each nonzero row is to the right of the leading entry of the row above it
- Each leading entry is the only nonzero entry in its column
For instance:
In echelon form after Gaussian Elimination becomes:
Antidiagonal matrix
Matrices are said to be antidiagonal when all the entries are zero but the antidiagonal (i.e., the diagonal starting from the bottom left corner to the upper right corner). For instance:
Design matrix
Design matrix is a special name for matrices containing explanatory variables or features in the context of statistics and machine learning. Some authors favor this name to refer to the set of variables or features in a model.
Matrices as systems of linear equations
I introduced the idea of systems of linear equations as a way to figure out the right combination of linear segments to obtain an outcome. I did this in the context of vectors, now we can extend this to the context of matrices.
Matrices are ideal to represent systems of linear equations. Consider the matrix and vectors and in . We can set up a system of linear equations as as:
This is equivalent to:
Geometrically, the solution for this representation equals to plot a set of planes in 3-dimensional space, one for each equation, and to find the segment where the planes intersect.
An alternative way, which I personally prefer to use, is to represent the system as a linear combination of the column vectors times a scaling term:
Geometrically, the solution for this representation equals to plot a set of vectors in 3-dimensional space, one for each column vector, then scale them by and add them up, tip to tail, to find the resulting vector .
The four fundamental matrix subsapces
Let's recall the definition of a subspace in the context of vectors:
- Contains the zero vector,
- Closure under multiplication,
- Closure under addition,
These conditions carry on to matrices since matrices are simply collections of vectors. Thus, now we can ask what are all possible subspaces that can be "covered" by a collection of vectors in a matrix. Turns out, there are four fundamental subspaces that can be "covered" by a matrix of valid vectors: (1) the column space, (2) the row space, (3) the null space, and (4) the left null space or null space of the transpose.
These subspaces are considered fundamental because they express many important properties of matrices in linear algebra.
The column space
The column space of a matrix is composed by all linear combinations of the columns of . We denote the column space as . In other words, equals to the span of the columns of . This view of a matrix is what we represented in Fig. 12: vectors in scaled by real numbers.
For a matrix and a vector , the column space is defined as:
In words: all linear combinations of the column vectors of and entries of an dimensional vector .
The row space
The row space of a matrix is composed of all linear combinations of the rows of a matrix. We denote the row space as . In other words, equals to the span of the rows of . Geometrically, this is the way we represented a matrix in Fig. 11: each row equation represented as planes. Now, a different way to see the row space, is by transposing . Now, we can define the row space simply as
For a matrix and a vector , the row space is defined as:
In words: all linear combinations of the row vectors of and entries of an dimensional vector .
The null space
The null space of a matrix is composed of all vectors that are map into the zero vector when multiplied by . We denote the null space as .
For a matrix and a vector , the null space is defined as:
The null space of the transpose
The left null space of a matrix is composed of all vectors that are map into the zero vector when multiplied by from the left. By "from the left", the vectors on the left of . We denote the left null space as
For a matrix and a vector , the null space is defined as:
Solving systems of linear equations with Matrices
Gaussian Elimination
When I was in high school, I learned to solve systems of two or three equations by the methods of elimination and substitution. Nevertheless, as systems of equations get larger and more complicated, such inspection-based methods become impractical. By inspection-based, I mean "just by looking at the equations and using common sense". Thus, to approach such kind of systems we can use the method of Gaussian Elimination.
Gaussian Elimination, is a robust algorithm to solve linear systems. We say is robust, because it works in general, it all possible circumstances. It works by eliminating terms from a system of equations, such that it is simplified to the point where we obtain the row echelon form of the matrix. A matrix is in row echelon form when all rows contain zeros at the bottom left of the matrix. For instance:
The values along the diagonal are the pivots also known as basic variables of the matrix. An important remark about the pivots, is that they indicate which vectors are linearly independent in the matrix, once the matrix has been reduced to the row echelon form.
There are three elementary transformations in Gaussian Elimination that when combined, allow simplifying any system to its row echelon form:
- Addition and subtraction of two equations (rows)
- Multiplication of an equation (rows) by a number
- Switching equations (rows)
Consider the following system :
We want to know what combination of columns of will generate the target vector . Alternatively, we can see this as a decomposition problem, as how can we decompose into columns of . To aid the application of Gaussian Elimination, we can generate an augmented matrix , this is, appending to on this manner:
We start by multiplying row 1 by and substracting it from row 2 as to obtain:
If we substract row 1 from row 3 as we get:
At this point, we have found the row echelon form of . If we divide row 3 by -3, We know that . By backsubsitution, we can solve for as:
Again, taking and we can solve for as:
In this manner, we have found that the solution for our system is , , and .
In NumPy, we can solve a system of equations with Gaussian Elimination with the linalg.solve method as:
A = np.array([[1, 3, 5],
[2, 2, -1],
[1, 3, 2]])
y = np.array([[-1],
[1],
[2]])
np.linalg.solve(A, y)
"""
array([[-2.],
[ 2.],
[-1.]])
"""
Which confirms our solution is correct.
Gauss-Jordan Elimination
The only difference between Gaussian Elimination and Gauss-Jordan Elimination, is that this time we "keep going" with the elemental row operations until we obtain the reduced row echelon form. The reduced part means two additionak things: (1) the pivots must be , (2) and the entries above the pivots must be . This is simplest form a system of linear equations can take. For instance, for a 3x3 matrix:
Let's retake from where we left Gaussian elimination in the above section. If we divide row 3 by -3 and row 2 by -4 as and , we get:
Again, by this point we we know . If we multiply row 2 by 3 and substract from row 1 as :
Finally, we can add 3.25 times row 3 to row 1, and substract 2.75 times row 3 to row 2, as and to get the reduced row echelon form as:
Now, by simply following the rules of matrix-vector multiplication, we get =
There you go, we obtained that , , and .
Matrix basis and rank
A set of linearly independent column vectors with elements forms a basis. For instance, the column vectors of are a basis:
"A basis for what?" You may be wondering. In the case of , for any vector . On the contrary, the column vectors for do not form a basis for :
In the case of , the third column vector is a linear combination of first and second column vectors.
The definition of a basis depends on the independence-dimension inequality, which states that a linearly independent set of vectors can have at most elements. Alternatively, we say that any set of vectors with elements is, necessarily, linearly dependent. Given that each vector in a basis is linearly independent, we say that any vector with elements, can be generated in a unique linear combination of the basis vectors. Hence, any matrix more columns than rows (as in ) will have dependent vectors. Basis are sometimes referred to as the minimal generating set.
An important question is how to find the basis for a matrix. Another way to put the same question is to found out which vectors are linearly independent of each other. Hence, we need to solve:
Where are the column vectors of . We can approach this by using Gaussian Elimination or Gauss-Jordan Elimination and reducing to its row echelon form or reduced row echelon form. In either case, recall that the pivots of the echelon form indicate the set of linearly independent vectors in a matrix.
NumPy does not have a method to obtain the row echelon form of a matrix. But, we can use Sympy, a Python library for symbolic mathematics that counts with a module for Matrices operations.SymPy has a method to obtain the reduced row echelon form and the pivots, rref.
from sympy import Matrix
A = Matrix([[1, 0, 1],
[0, 1, 1]])
B = Matrix([[1, 2, 3, -1],
[2, -1, -4, 8],
[-1, 1, 3, -5],
[-1, 2, 5, -6],
[-1, -2, -3, 1]])
A_rref, A_pivots = A.rref()
print('Reduced row echelon form of A:')
# Reduced row echelon form of A:
print(f'Column pivots of A: {A_pivots}')
# Column pivots of A: (0, 1)
B_rref, B_pivots = B.rref()
print('Reduced row echelon form of B:')
# Reduced row echelon form of B:
print(f'Column pivots of A: {B_pivots}')
# Column pivots of A: (0, 1, 3)
For , we found that the first and second column vectors are the basis, whereas for is the first, second, and fourth.
Now that we know about a basis and how to find it, understanding the concept of rank is simpler. The rank of a matrix is the dimensionality of the vector space generated by its number of linearly independent column vectors. This happens to be identical to the dimensionality of the vector space generated by its row vectors. We denote the rank of matrix as or .
For an square matrix (i.e., ), we say is full rank when every column and/or row is linearly independent. For a non-square matrix with (i.e., more rows than columns), we say is full rank when every row is linearly independent. When (i.e., more columns than rows), we say is full rank when every column is linearly independent.
From an applied machine learning perspective, the rank of a matrix is relevant as a measure of the information content of the matrix. Take matrix from the example above. Although the original matrix has 5 columns, we know is rank 4, hence, it has less information than it appears at first glance.
Matrix norm
As with vectors, we can measure the size of a matrix by computing its norm. There are multiple ways to define the norm for a matrix, as long it satisfies the same properties defined for vectors norms: (1) absolutely homogeneous, (2) triangle inequality, (3) positive definite (see vector norms section). For our purposes, I'll cover two of the most commonly used norms in machine learning: (1) Frobenius norm, (2) max norm, (3) spectral norm.
Note: I won't cover the spectral norm just yet, because it depends on concepts that I have not introduced at this point.
Frobenius norm
The Frobenius norm is an element-wise norm named after the German mathematician Ferdinand Georg Frobenius. We denote this norm as . You can thing about this norm as flattening out the matrix into a long vector. For instance, a matrix would become a vector with entries. We define the Frobenius norm as:
In words: square each entry of , add them together, and then take the square root.
In NumPy, we can compute the Frobenius norm as with the linal.norm method ant fro as the argument:
A = np.array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
np.linalg.norm(A, 'fro')
# 16.881943016134134
Max norm
The max norm or infinity norm of a matrix equals to the largest sum of the absolute value of row vectors. We denote the max norm as . Consider . We define the max norm for as:
This equals to go row by row, adding the absolute value of each entry, and then selecting the largest sum.
In Numpy, we compute the max norm as:
A = np.array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
np.linalg.norm(A, np.inf)
# 24.0
In this case, is easy to see that the third row has the largest absolute value.
Spectral norm
To understand this norm, is necessary to first learn about eigenvectors and eigenvalues, which I cover later.
The spectral norm of a matrix equals to the largest singular value . We denote the spectral norm as . Consider . We define the spectral for as:
In Numpy, we compute the max norm as:
A = np.array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9]])
np.linalg.norm(A, 2)
# 16.84810335261421
Linear and affine mappings
# Libraries for this section
import numpy as np
import pandas as pd
import altair as alt
alt.themes.enable('dark')
# ThemeRegistry.enable('dark')
Linear mappings
Now we have covered the basics of vectors and matrices, we are ready to introduce the idea of a linear mapping. Linear mappings, also known as linear transformations and linear functions, indicate the correspondence between vectors in a vector space and the same vectors in a different vector space . This is an abstract idea. I like to think about this in the following manner: imagine there is a multiverse as in Marvel comics, but instead of humans, aliens, gods, stars, galaxies, and superheroes, we have vectors. In this context, a linear mapping would indicate the correspondence of entities (i.e., planets, humans, superheroes, etc) between universes. Just imagine us, placidly existing in our own universe, and suddenly a linear mapping happens: our entire universe would be transformed into a different one, according to whatever rules the linear mapping has enforced. Now, switch universes for vector spaces and us by vectors, and you'll get the full picture.
So, linear mappings transform vector spaces into others. Yet, such transformations are constrained to a spefic kind: linear ones. Consider a linear mapping and a pair of vectors and . To be valid, a linear mapping must satisfies these rules:
In words:
- The transformation of the sum of the vectors must be equal to taking the transformation of each vector individually and then adding them up.
- The transformation of a scaled version of a vector must be equal to taking the transformation of the vector first and then scaling the result.
The two properties above can be condenced into one, the superposition property:
As a result of satisfying those properties, linear mappings preserve the structure of the original vector space. Imagine a vector space , like a grid on lines in a cartesian plane. Visually, preserving the structure of the vector space after a mapping means to: (1) the origin of the coordinate space remains fixed, and (2) the lines remain lines and parallel to each other.
In linear algebra, linear mappings are represented as matrices and performed by matrix multiplication. Take a vector and a matrix . We say that when multiplies , the matrix transform the vector into another one:
The typicall notation for a linear mapping is the same we used for functions. For the vector spaces and , we indicate the linear mapping as
Examples of linear mappings
Let's examine a couple of examples of proper linear mappings. In general, dot products are linear mappings. This should come as no surprise since dot products are linear operations by definition. Dot products sometimes take special names, when they have a well-known effect on a linear space. I'll examine two simple cases: negation and reversal. Keep in mind that although we will test this for one vector, this mapping work on the entire vector space (i.e., the span) of a given dimensionality.
Negation matrix
A negation matrix returns the opposite sign of each element of a vector. It can be defined as:
This is, the negative identity matrix. Consider a pair of vectors and , and the negation matrix . Let's test the linear mapping properties with NumPy:
x = np.array([[-1],
[0],
[1]])
y = np.array([[-3],
[0],
[2]])
T = np.array([[-1,0,0],
[0,-1,0],
[0,0,-1]])
We first test :
left_side_1 = T @ (x+y)
right_side_1 = (T @ x) + (T @ y)
print(f"Left side of the equation:\n{left_side_1}")
print(f"Right side of the equation:\n{right_side_1}")
"""
Left side of the equation:
[[ 4]
[ 0]
[-3]]
Right side of the equation:
[[ 4]
[ 0]
[-3]]
"""
Hence, we confirm we get the same results.
Let's check the second property
alpha = 2
left_side_2 = T @ (alpha * x)
right_side_2 = alpha * (T @ x)
print(f"Left side of the equation:\n{left_side_2}")
print(f"Right side of the equation:\n{right_side_2}")
"""
Left side of the equation:
[[ 2]
[ 0]
[-2]]
Right side of the equation:
[[ 2]
[ 0]
[-2]]
"""
Again, we confirm we get the same results for both sides of the equation
Reversal matrix
A reversal matrix returns reverses the order of the elements of a vector. This is, the last become the first, the second to last becomes the second, and so on. For a matrix in is defined as:
In general, it is the identity matrix but backwards, with ones from the bottom left corner to the top right corern. Consider a pair of vectors and , and the reversal matrix . Let's test the linear mapping properties with NumPy:
x = np.array([[-1],
[0],
[1]])
y = np.array([[-3],
[0],
[2]])
T = np.array([[0,0,1],
[0,1,0],
[1,0,0]])
We first test :
x_reversal = T @ x
y_reversal = T @ y
left_side_1 = T @ (x+y)
right_side_1 = (T @ x) + (T @ y)
print(f"x before reversal:\n{x}\nx after reversal \n{x_reversal}")
print(f"y before reversal:\n{y}\ny after reversal \n{y_reversal}")
print(f"Left side of the equation (add reversed vectors):\n{left_side_1}")
print(f"Right side of the equation (add reversed vectors):\n{right_side_1}")
"""
x before reversal:
[[-1]
[ 0]
[ 1]]
x after reversal
[[ 1]
[ 0]
[-1]]
y before reversal:
[[-3]
[ 0]
[ 2]]
y after reversal
[[ 2]
[ 0]
[-3]]
Left side of the equation (add reversed vectors):
[[ 3]
[ 0]
[-4]]
Right side of the equation (add reversed vectors):
[[ 3]
[ 0]
[-4]]
"""
This works fine. Let's check the second property
alpha = 2
left_side_2 = T @ (alpha * x)
right_side_2 = alpha * (T @ x)
print(f"Left side of the equation:\n{left_side_2}")
print(f"Right side of the equation:\n{right_side_2}")
"""
Left side of the equation:
[[ 2]
[ 0]
[-2]]
Right side of the equation:
[[ 2]
[ 0]
[-2]]
"""
Examples of nonlinear mappings
As with most subjects, examining examples of what things are not can be enlightening. Let's take a couple of non-linear mappings: norms and translation.
Norms
This may come as a surprise, but norms are not linear transformations. Not "some" norms, but all norms. This is because of the very definition of a norm, in particular, the triangle inequality and positive definite properties, colliding with the requirements of linear mappings.
First, the triangle inequality defines: . Whereas the first requirement for linear mappings demands: . The problem here is in the condition, which means adding two vectors and then taking the norm can be less than the sum of the norms of the individual vectors. Such condition is, by defnition, not allowed for linear mappings.
Second, the positive definite defines: and . Put simply, norms have to be a postive value. For instance, the norm of , instead of . But, the second property for linear mappings requires . Hence, it fails when we multiply by a negative number (i.e., it can preserve the negative sign).
Translation
Translation is a geometric transformation that moves every vector in a vector space by the same distance in a given direction. Translation is an operation that matches our everyday life intuitions: move a cup of coffee from your left to your right, and you would have performed translation in space.
Contrary to what we have seen so far, the translation matrix is represented with homogeneous coordinates instead of cartesian coordinates. Put simply, the homogeneous coordinate system adds a extra at the end of vectros. For instance, the vector in cartesian coordinates:
Becomes the following in homogeneous coordinates:
In fact, the translation matrix for the general case can't be represented with cartesian coordinates. Homogeneous coordinates are the standard in fields like computer graphics since they allow us to better represent a series of transformations (or mappings) like scaling, translation, rotation, etc.
A translation matrix in can be denoted as:
Where and are the values added to each dimension for translation. For instance, consider . If we want translate this units in the first dimension, and units in the second dimension, we first transfor the vector to homogeneous coordinates , and then perfom matrix-vector multiplication as usual:
The first two vectors in the translation matrix simple reproduce the original vector (i.e., the identity), and the third vector is the one actually "moving" the vectors.
Translation is not a linear mapping simply because does not hold. In the case of translation , which invalidates the operation as a linear mapping. This type of mapping is known as an affine mapping or transformation, which is the topic I'll review next.
Affine mappings
The simplest way to describe affine mappings (or transformations) is as a linear mapping + translation. Hence, an affine mapping takes the form of:
Where is a linear mapping or transformation and is the translation vector.
If you are familiar with linear regression, you would notice that the above expression is its matrix form. Linear regression is usually analyzed as a linear mapping plus noise, but it can also be seen as an affine mapping. Alternative, we can say that is a linear mapping if and only if .
From a geometrical perspective, affine mappings displace spaces (lines or hyperplanes) from the origin of the coordinate space. Consequently, affine mappings do not operate over vector spaces as the zero vector condition does not hold anymore. Affine mappings act onto affine subspaces, that I'll define later in this section.
Affine combination of vectors
We can think in affine combinations of vectors, as linear combinations with an added constraint. Let's recall de definitoon for a linear combination. Consider a set of vectors and scalars , then a linear combination is:
For affine combinations, we add the condition:
In words, we constrain the sum of the weights to . In practice, this defines a weighted average of the vectors. This restriction has a palpable effect which is easier to grasp from a geometric perspective.
Fig. 15 shows two affine combinations. The first combination with weights and , which yields the midpoint between vectors and . The second combination with weights and (add up to ), which yield a point over the vector . In both cases, we have that the resulting vector lies on the same line. This is a general consequence of constraining the sum of the weights to : every affine combination of the same set of vectors will map onto the same space.
Affine span
The set of all linear combinations, define the the vector span. Similarly, the set of all affine combinations determine the affine span. As we saw in Fig. 15, every affine of vectors and maps onto the line . More generally, we say that the affine span of vectors is:
Again, in words: the affine span is the set of all linear combinations of the vector set, such that the weights add up to and all weights are real numbers. Hence, the fundamental difference between vector spaces and affine spaces, is the former will span the entire space (assuming independent vectors), whereas the latter will span a line.
Let's consider three cases in : (1) three linearly independent vectors; (2) two linearly independent vectors and one dependent vector; (3) three linearly dependent vectors. In case (1), the affine span is the 2-dimensional plane containing those vectors. In case (2), the affine space is a line. Finally, in case (3), the span a single point. This may not be entirely obvious, so I encourage you to draw and the three cases, take the affine combinations and see what happens.
Affine space and subspace
In simple terms, affine spaces are translates of vector spaces, this is, vector spaces that have been offset from the origin of the coordinate system. Such a notion makes sound affine spaces as a special case of vector spaces, but they are actually more general. Indeed, affine spaces provide a more general framework to do geometric manipulation, as they work independently of the choice of the coordinate system (i.e., it is not constrained to the origin). For instance, the set of solutions of the system of linear equations (i.e., linear regression), is an affine space, not a linear vector space.
Consider a vector space , a vector , and a subset . We define an affine subspace as:
Further, any point, line, plane, or hyperplane in that does not go through the origin, is an affine subspace.
Affine mappings using the augmented matrix
Consider the matrix , and vectors
We can represent the system of linear equations:
As a single matrix vector multiplication, by using an augmented matrix of the form:
This form is known as the affine transformation matrix. We made use of this form when we exemplified translation, which happens to be an affine mapping.
Special linear mappings
There are several important linear mappings (or transformations) that can be expressed as matrix-vector multiplications of the form . Such mappings are common in image processing, computer vision, and other linear applications. Further, combinations of linear and nonlinear mappings are what complex models as neural networks do to learn mappings from inputs to outputs. Here we briefly review six of the most important linear mappings.
Scaling
Scaling is a mapping of the form , with . Scaling stretches by a factor when , shrinks when , and reverses the direction of the vector when . For geometrical objects in Euclidean space, scaling changes the size but not the shape of objects. An scaling matrix in takes the form:
Where are the scaling factors.
Let's scale a vector using NumPy. We will define a scaling matrix , a vector to scale, and then plot the original and scaled vectors with Altair.
A = np.array([[2.0, 0],
[0, 2.0]])
x = np.array([[0, 2.0,],
[0, 4.0,]])
To scale , we perform matrix-vector multiplication as usual
y = A @ x
z = np.column_stack((y,x))
df = pd.DataFrame({'dim-1': z[0], 'dim-2':z[1], 'type': ['tran', 'tran', 'base', 'base']})
df
| dim-1 | dim-2 | type | |
|---|---|---|---|
| 0 | 0.0 | 0.0 | tran |
| 1 | 4.0 | 8.0 | tran |
| 2 | 0.0 | 0.0 | base |
| 3 | 2.0 | 4.0 | base |
We see that the resulting scaled vector ('tran') is indeed two times the original vector ('base'). Now let's plot. The light blue line solid line represents the original vector, whereas the dashed orange line represents the scaled vector.
chart = alt.Chart(df).mark_line(opacity=0.8).encode(
x='dim-1',
y='dim-2',
color='type',
strokeDash='type')
chart
Reflection
Reflection is the mirror image of an object in Euclidean space. For the general case, reflection of a vector through a line that passes through the origin is obtained as:
where are radians of inclination with respect to the horizontal axis. I've been purposely avoiding trigonometric functions, so let's examine a couple of special cases for a vector in (that can be extended to an arbitrary number of dimensions).
Reflection along the horizontal axis, or around the line at from the origin:
Reflection along the vertical axis, or around the line at from the origin:
Reflection along the line where the horizontal axis equals the vertical axis, or around the line at from the origin:
Reflection along the line where the horizontal axis equals the negative of the vertical axis, or around the line at from the origin:
Let's reflect a vector using NumPy. We will define a reflection matrix , a vector to reflect, and then plot the original and reflected vectors with Altair.
# rotation along the horiontal axis
A1 = np.array([[1.0, 0],
[0, -1.0]])
# rotation along the vertical axis
A2 = np.array([[-1.0, 0],
[0, 1.0]])
# rotation along the line at 45 degrees from the origin
A3 = np.array([[0, 1.0],
[1.0, 0]])
# rotation along the line at -45 degrees from the origin
A4 = np.array([[0, -1.0],
[-1.0, 0]])
x = np.array([[0, 2.0,],
[0, 4.0,]])
y1 = A1 @ x
y2 = A2 @ x
y3 = A3 @ x
y4 = A4 @ x
z = np.column_stack((x, y1, y2, y3, y4))
df = pd.DataFrame({'dim-1': z[0], 'dim-2':z[1],
'reflection': ['original', 'original',
'0-degrees', '0-degrees',
'90-degrees', '90-degrees',
'45-degrees', '45-degrees',
'neg-45-degrees', 'neg-45-degrees']})
df
| dim-1 | dim-2 | reflection | |
|---|---|---|---|
| 0 | 0.0 | 0.0 | original |
| 1 | 2.0 | 4.0 | original |
| 2 | 0.0 | 0.0 | 0-degrees |
| 3 | 2.0 | -4.0 | 0-degrees |
| 4 | 0.0 | 0.0 | 90-degrees |
| 5 | -2.0 | 4.0 | 90-degrees |
| 6 | 0.0 | 0.0 | 45-degrees |
| 7 | 4.0 | 2.0 | 45-degrees |
| 8 | 0.0 | 0.0 | neg-45-degrees |
| 9 | -4.0 | -2.0 | neg-45-degrees |
def base_coor(ran1: float, ran2: float):
'''return base chart with coordinate space'''
df_base = pd.DataFrame({'horizontal': np.linspace(ran1, ran2, num=2), 'vertical': np.zeros(2)})
h = alt.Chart(df_base).mark_line(color='white').encode(
x='horizontal',
y='vertical')
v = alt.Chart(df_base).mark_line(color='white').encode(
y='horizontal',
x='vertical')
base = h + v
return base
chart = alt.Chart(df).mark_line().encode(
x=alt.X('dim-1', axis=alt.Axis(title='horizontal-axis')),
y=alt.Y('dim-2', axis=alt.Axis(title='vertical-axis')),
color='reflection')
base_coor(-5.0, 5.0) + chart
Shear
Shear mappings are hard to describe in words but easy to understand with images. I recommend to look at the shear mapping below and then read this description: a shear mapping displaces points of an object in a given direction (e.g., all points to the right), in a proportion equal to their perpendicular distance from an axis (e.g., the line on the axis) that remains fixed. A "proportion equal to their perpendicular distance" means that points further away from the reference axis displace more than points near to the axis.
For an object in , a horizontal shear matrix (i.e., paraller to the horizontal axis) takes the form:
Where is the shear factor, that essentially determines how pronounced is the shear.
For an object in , a vertical shear matrix (i.e., paraller to the vertical axis) takes the form:
Let's shear a vector using NumPy. We will define a shear matrix , a pair of vectors and to shear, and then plot the original and shear vectors with Altair. The reason we define two vectors, is that shear mappings are easier to appreciate with planes or multiple sides figures than single lines.
# shear along the horiontal axis
A1 = np.array([[1.0, 1.5],
[0, 1.0]])
x = np.array([[0, 2.0,],
[0, 4.0,]])
u = np.array([[2, 4.0,],
[0, 4.0,]])
y1 = A1 @ x
v1 = A1 @ u
z = np.column_stack((x, y1, u, v1))
df = pd.DataFrame({'dim-1': z[0], 'dim-2':z[1],
'shear': ['original', 'original',
'horizontal', 'horizontal',
'original-2', 'original-2',
'horizontal-2', 'horizontal-2'
]})
chart = alt.Chart(df).mark_line().encode(
x=alt.X('dim-1', axis=alt.Axis(title='horizontal-axis')),
y=alt.Y('dim-2', axis=alt.Axis(title='vertical-axis')),
color='shear')
base_coor(-5.0, 10.0) + chart
Rotation
Rotation mappings do exactly what their name indicates: they move objects (by convection) counterclockwise in Euclidean space. For the general case in , counterclockwise of vector by radiants rotations is obtained as:
Again, let's examine a couple of special cases.
A rotation matrix in :
A rotation matrix in :
A rotation matrix in :
Let's rotate a vector using NumPy. We will define a rotation matrix , a vector , and then plot the original and rotated vectors with Altair.
# 90-degrees roration
A1 = np.array([[0, -1.0],
[1, 0]])
# 180-degrees roration
A2 = np.array([[-1.0, 0],
[0, -1.0]])
# 270-degrees roration
A3 = np.array([[0, 1.0],
[-1.0, 0]])
x = np.array([[0, 2.0,],
[0, 4.0,]])
y1 = A1 @ x
y2 = A2 @ x
y3 = A3 @ x
z = np.column_stack((x, y1, y2, y3))
df = pd.DataFrame({'dim-1': z[0], 'dim-2':z[1],
'rotation': ['original', 'original',
'90-degrees', '90-degrees',
'180-degrees', '180-degrees',
'270-degrees', '270-degrees'
]})
df
| dim-1 | dim-2 | rotation | |
|---|---|---|---|
| 0 | 0.0 | 0.0 | original |
| 1 | 2.0 | 4.0 | original |
| 2 | 0.0 | 0.0 | 90-degrees |
| 3 | -4.0 | 2.0 | 90-degrees |
| 4 | 0.0 | 0.0 | 180-degrees |
| 5 | -2.0 | -4.0 | 180-degrees |
| 6 | 0.0 | 0.0 | 270-degrees |
| 7 | 4.0 | -2.0 | 270-degrees |
chart = alt.Chart(df).mark_line().encode(
x=alt.X('dim-1', axis=alt.Axis(title='horizontal-axis')),
y=alt.Y('dim-2', axis=alt.Axis(title='vertical-axis')),
color='rotation')
base_coor(-5.0, 5.0) + chart
Projections
Projections are a fundamental type of linear (and affine) mappings for machine learning. If you have ever heard concepts like "embeddings", "low-dimensional representation", or "dimensionality reduction", they all are examples of projections. Even linear regression and principal component analysis are exemplars of projections. Thus, projections allow working with high-dimensional spaces (i.e., problems with many features or variables) more efficiently, by projecting such spaces into lower-dimensional spaces. this works because is often the case that a few dimensions contain most of the information to understand the relation between inputs and outputs. Moreover, projections can be represented as matrices acting on vectors.
Put simply, projections are mappings from a space onto a subpace, or from a set of vectors onto a subset of vectors. Additionally, projections are "idempotent", this is, the projection has the property to be equal to its composition with itself. In other words, when you wrap a projection into itself as , the result does not change, i.e., . Formally, for a vector space and a vector subset , we define a projection as:
with
Here we are concerned with the matrix representation of projections, which receive the special name of projection matrices, denoted as . By extension, projection matrices are also "idempotent":
Projections onto lines
In Freudian psychoanalysis, projection is a defense mechanism of the "ego" (i.e., the sense of self), where a person denies the possession of an undesired characteristic while attributing it to someone else, i.e., "projecting" what we don't like of us onto others.
It turns out, that the concept of projection in mathematics is not that different from the Freudian one. Just make the following analogy: imagine you and foe of you are represented as vectors in a 2-dimensional cartesian plane, as and respectively. The way on which you would project yourself onto your foe is by tracing a perpendicular line () from you onto him. Why perpendicular? Because this is the shortest distance between you and him, hence, the most efficient way to project yourself onto him. Now, the projection would be "how much" of yourself was "splattered" onto him, which is represented by the segment from the origin until the point where the perpendicular line touched your foe.
Now, recall that lines crossing the origin form subspaces, hence vector is a subspace, and that perpendicular lines form angles, hence the projection is orthogonal. More formally, we can define the projection of onto subspace formed by as:
Where must be the minimal distance between and (i.e., and ), where distance is:
Further, the resulting projection must lie in the span of . Therefore, we can conclude that , where alpha is a scalar in .
The formula to find the orthogonal projection (I'm skipping the derivation on purpose) is:
In words: we take the dot product between and , divide by the norm of , and multiply by . In this case, is also known as a basis vector, so we can say that is projected onto the basis .
Now, we want to express projections as matrices, i.e., as the matrix vector product . For this, recall that matrix-scalar multiplication is commutative, hence we can perform a little of algeabric manipulation to find:
In this form, we can indeed express the projection as a matrix-vector multiplication, because results in a symmetrix matrix, and is a scalar, which means that it can be expressed as a matrix:
In sum, the matrix will project any vector onto .
Let's use NumPy to find the projection from onto a basis vector .
# base vector
y = np.array([[3],
[2]])
x = np.array([[1],
[3]])
P = (y @ y.T)/(y.T @ y)
print(f'Projection matrix for y:\n{P}')
"""
Projection matrix for y:
[[0.69230769 0.46153846]
[0.46153846 0.30769231]]
"""
z = P @ x
print(f'Projection from x onto y:\n{z}')
"""
Projection from x onto y:
[[2.07692308]
[1.38461538]]
"""
Let's plot the vectors to make things clearer
# origin coordinate space
o = np.array([[0],
[0]])
v = np.column_stack((o, x, o, y, o, z, x, z))
df = pd.DataFrame({'dim-1': v[0], 'dim-2':v[1],
'vector': ['x-vector', 'x-vector',
'y-base-vector', 'y-base-vector',
'z-projection', 'z-projection',
'orthogonal-vector', 'orthogonal-vector'],
'size-line': [2, 2, 2, 2, 4, 4, 2, 2]})
df
| dim-1 | dim-2 | vector | size-line | |
|---|---|---|---|---|
| 0 | 0.000000 | 0.000000 | x-vector | 2 |
| 1 | 1.000000 | 3.000000 | x-vector | 2 |
| 2 | 0.000000 | 0.000000 | y-base-vector | 2 |
| 3 | 3.000000 | 2.000000 | y-base-vector | 2 |
| 4 | 0.000000 | 0.000000 | z-projection | 4 |
| 5 | 2.076923 | 1.384615 | z-projection | 4 |
| 6 | 1.000000 | 3.000000 | orthogonal-vector | 2 |
| 7 | 2.076923 | 1.384615 | orthogonal-vector | 2 |
chart = alt.Chart(df).mark_line().encode(
x=alt.X('dim-1', axis=alt.Axis(title='horizontal-axis')),
y=alt.Y('dim-2', axis=alt.Axis(title='vertical-axis')),
color='vector',
strokeDash='vector',
size = 'size-line')
base_coor(-1.0, 4.0) + chart
Projections onto general subspaces
From the previous section, we learned that the projection matrix for the one-dimensional project of onto (i.e. ) can be expressed as:
Which implies that we the projection is entirely defined in terms of the basis subspace. Now, we are interested in projections for the general case, this is, for set of basis vectors . By extension, we can define such projection as:
Where is the matrix of basis vectors.
Nothing fancy going on here: we just need to take the sum for the product between each basis vector and . As with the one-dimensional case, we want the projection to be the minimal distance from onto , which we know implies orthogonal lines (or hyperplanes) connecting with . The condition for orthogonality (again, I'm skipping the derivation on purpose) here equals:
Now, recall what we really want is to find (we know already). Therefore, with a bit of algeabric manipulation we can clear the expression above as:
Such expression is known as the pseudo-inverse or Moore–Penrose inverse of . To work, it requires to be full rank (i.e., independent columns, which should be the case for basis). It can be used to solve linear regression problems, although you'll probably find the notation flipped as: (my bad choice of notation!).
Going back to our Freudian projection analogy, this is like a group of people projecting themselves onto someone else, with that person representing a rough approximation of the character of the group.
Projections as approximate solutions to systems of linear equations
Machine learning prediction problems usually require to find a solution to systems of linear equations of the form:
In other words, to represent as linear combinations of the columns of . Unfortunately, in most cases is not in the column space of , i.e., there is no way to find a linear combination of its columns to obtain the target . In such cases, we can use orthogonal projections to find approximate solutions to the system. We usually denote approximated solutions for systems of linear equations as . Now, will be in the span of the columns of and will be the result of projecting onto the subspace of the columns of . That solution will be the best (closest) approximation of given the span of the columns of . In sum: the approximated solution is the orthogonal projection of onto .
Matrix decompositions
In the Japanese manga/anime series Fullmetal Alchemist, Alchemy is understood as the metaphysical science of altering objects by manipulating its natural components, act known as Transmutation (Rensei). There are three steps to Transmutation: (1) Comprehension, to understand the atomic structure and properties of the object, (2) Deconstruction, to break down the structure of the object into its fundamental elements (3) Reconstruction, to use the natural flow of energy to reform the object into a new shape.
Metaphorically speaking, we can understand linear combinations and matrix decompositions in analogy to Transmutation. Matrix decomposition is essentially about to break down a matrix into simpler "elements" or matrices (deconstruction), which allows us to better understand its fundamental structure (comprehension). Linear combinations are essentially about taking the fundamental elements of a matrix (i.e., set of vectors) to generate a new object.
Matrix decomposition is also known as matrix factorization, in reference the fact that matrices can be broken down into simpler matrices, more on less in the same way that Prime factorization breaks down large numbers into simpler primes (e.g., ).
There are several important applications of matrix factorization in machine learning: clustering, recommender systems, dimensionality reduction, topic modeling, and others. In what follows I'll cover a selection of several basic and common matrix decomposition techniques.
LU decomposition
There are multiple ways to decompose or factorize matrices. One of the simplest ways is by decomposition a matrix into a lower triangular matrix and an upper triangular matrix, the so-called LU or Lower-Upper decomposition.
LU decomposition is of great interest to us since it's one of the methods computers use to solve linear algebra problems. In particular, LU decomposition is a way to represent Gaussian Elimination in numerical linear algebra. LU decomposition is flexible as it can be obtained from noninvertible or singular matrices, and from non-square matrices.
The general expression for LU decomposition is:
Meaning that can be represented as the product of the lower triangular matrix and upper triangular matrix . In the next sections, we explain the mechanics of the LU decomposition.
Elementary matrices
Our first step to approach LU decomposition is to introduce elementary matrices. When considering matrices as functions or mappings, we can associate special meaning to a couple of basic or "elementary" operations performed by matrices. Our starting point is the identity matrix, for instance:
As we saw before, the identity matrix does not change the values of another matrix under multiplication:
Because it is essentially saying: give me of each column of the matrix, i.e., return the original matrix. Now, consider the following matrix:
The only thing we did to the to obtain was to multiply the first row by . This can be considered an elementary operation. Here is another example:
Clearly, we can't obtain by multiplication only. From a column perspective, what we did was to add times the second column to the first column. Alternatively, from a row perspective, we can say we added times the first row to the second row.
One last example:
From the column perspective, we added times the third column to the second column. From the row perspective, we added times the second row to the third row.
You can probably see the pattern by now: by performing simple or "elementary" column or row operations, this is, multiplication and addition, we can obtain any lower triangular matrix. This type of matrices are what we call elementary matrices. In a way, we can say elementary matrices "encode" fundamental column and row operations. To see this, consider the following generic matrix:
Let's what happens when we multiply :
The result of reflects the same elementary operations we performed on to obtain from the column perspective: to add times the second column to the first one.
Now consider what happens when we multiply from the left:
Now we obtain the same elementary operations we performed on to obtain from the row perspective: to add times the first row to the second one.
The inverse of elementary matrices
A nice property of elementary matrices, is that the inverse is simply the opposite operation. For instance, the inverse of is:
This is because instead of multiplying the first row of by , we divide it by . Similarly, the inverse of is:
Again, instead of adding , we add (or substract ). Finally, the inverse of is:
The reason we care about elementary matrices and its inverse is that it will be fundamental to understand LU decomposition.
LU decomposition as Gaussian Elimination
Let's briefly recall Gaussian Elimination: it's an robust algorithm to solve systems of linear equations, by sequentially applying three elementary transformations:
- Addition and subtraction of two equations (rows)
- Multiplication of an equation (rows) by a number
- Switching equations (rows)
Gaussian Elimination will reduce matrices to its row echelon form, which is an upper triangular matrix, with zero rows at the bottom, and zeros below the pivot for each column.
It turns out, there is a clever way to organize the steps from Gaussian Elimination: with elementary matrices.
Consider the following matrix :
The first step consist of substracting two times row 1 from row 1. Before, we represented this operation as , and write down the result, which is:
Alternatively, as we learned in the previous section, we can represent row operations as multiplication by elementary matrices, to obtain the same result. Since we want to substract times the first row from the second, we need to (1) multiply from the left, and (2) add a to the first element of the second row:
You don't have to believe me. Let's confirm this is correct with NumPy:
A = np.array([[1, 3, 5],
[2, 2, -1],
[1, 3, 2]])
l1 = np.array([[1, 0, 0],
[-2, 1, 0],
[0, 0, 1]])
l1 @ A
"""
array([[ 1, 3, 5],
[ 0, -4, -11],
[ 1, 3, 2]])
"""
As you can see, the result is exactly what we obtained before by . But, we are not done. We still need to get rid of the and in the third row. For this, we would normally do to obtain:
Again, we can encode this using elementary matrices as:
Once again, let's confirm this with NumPy:
A = np.array([[1, 3, 5],
[2, 2, -1],
[1, 3, 2]])
l2 = np.array([[1, 0, 0],
[-2, 1, 0],
[-1, 0, 1]])
l2 @ A
"""
array([[ 1, 3, 5],
[ 0, -4, -11],
[ 0, 0, -3]])
"""
Indeed, the result is correct. At this point, we have reduced to its row echelon form. We will call to the resulting matrix from , as it is an upper triangular matrix. Hence, we arrived to the identity:
This is not quite LU decomposition. To get there, we just need to multiply both sides of the equality by the inverse of , that we will call , which yields:
There you go: we arrived to the LU decomposition expression. As a final note, recall that the inverse of is:
Let's confirm with NumPy this works, by multiplying by :
# inverse of l
L = np.array([[1, 0, 0],
[2, 1, 0],
[1, 0, 1]])
# upper triangular resulting from Gaussian Elimination
U = np.array([[1, 3, 5],
[0, -4, -11],
[0, 0, -3]])
L @ U
"""
array([[ 1, 3, 5],
[ 2, 2, -1],
[ 1, 3, 2]])
""""
Indeed, we recover by multiplying .
LU decomposition with pivoting
If you recall the three elementary operations allowed in Gaussian Elimination, we had: (1) multiplication, (2) addition, (3) switching. At this point, we haven't seen switching with LU decomposition. It turns out, that LU decomposition does not work when switching or permutations of rows are required to solve a system of linear equations. Further, even when pivoting is not required to solve a system, the numerical stability of Gaussian Elimination when implemented in computers is problematic, and pivoting helps to tackle that issue as well.
Let's see a simple example of pivoting. Consider the following matrix :
In this case, we can't get rid of the first in the second column by substraction. If we do that, we obtain:
Which is the opposite of what we want. A simple way to fix this is by switching rows 1 and 2 as:
And then substracting row 1 from row 2 to obtain:
Bam! Problem fixed. Now, as with multiplication and addition, we can represent permutations with matrices as well. In particular, by using permutation matrices. For our previous example, we can do:
Let's confirm this is correct with NumPy
P = np.array([[0, 1],
[1, 0]])
A = np.array([[0, 1],
[1, 1]])
P @ A
"""
array([[1, 1],
[0, 1]])
"""
It works. Now we can put all the pieces together and decompose by using the following expression:
This is known as LU decomposition with pivoting. An alternative expression of the same decomposition is:
In Python, we can use SciPy to perform LUP decomposition by using the linalg.lu method. Let's decompose a larger matrix to make things more interesting.
from scipy.linalg import lu
A = np.array([[2, 1, 1, 0],
[4, 3, 3, 1],
[8, 7, 9, 5],
[6, 7, 9, 8]])
P, L, U = lu(A)
print(f'Pivot matrix:\n{P}')
"""
Pivot matrix:
[[0. 0. 0. 1.]
[0. 0. 1. 0.]
[1. 0. 0. 0.]
[0. 1. 0. 0.]]
"""
print(f'Lower triangular matrix:\n{np.round(L, 2)}')
"""
Lower triangular matrix:
[[ 1. 0. 0. 0. ]
[ 0.75 1. 0. 0. ]
[ 0.5 -0.29 1. 0. ]
[ 0.25 -0.43 0.33 1. ]]
"""
print(f'Upper triangular matrix:\n{np.round(U, 2)}')
"""
Upper triangular matrix:
[[ 8. 7. 9. 5. ]
[ 0. 1.75 2.25 4.25]
[ 0. 0. -0.86 -0.29]
[ 0. 0. 0. 0.67]]
"""
We can confirm the decomposition is correct by multiplying the obtained matrices
A_recover = np.round(P @ L @ U, 1)
print(f'PLU multiplicatin:\n{A_recover.astype(int)}')
"""
PLU multiplicatin:
[[2 1 1 0]
[4 3 3 1]
[8 7 9 5]
[6 7 9 8]]
"""
We recover perfectly.
QR decomposition
QR decomposition or QR factorization, is another very relevant decomposition in the context of numerical linear algebra. As with LU decomposition, It can be used to solve systems of linear equations like least square problems and to find eigenvalues of a general matrix.
QR decomposition works by decomposing into an orthogonal matrix , and a upper traingular matrix as:
Next, we will review a few concepts to properly explain QR decomposition.
Orthonormal basis
In previous sections we learned about basis and orthogonal basis. Specifically, we said that a set of linearly independent column vectors with elements forms a basis. We also said that a pair of vectors and are orthogonal if their inner product is zero, or . Consequently, a set of orthogonal vectors form an orthogonal basis for a matrix and for the vector space spanned by such matrix.
To go from orthogonal basis vectors to orthonomal basis vectors, we just need to divide each vector by its lenght or norm. When we divide a basis vector by its norm we obtain a unit basis vector. More formally, a set of vectors is orthonormal if:
In words: when we take the inner product of a pair of orthogonal vectors, it results in ; when we take the inner product of a vector with itself, it results in .
For instance, consider and :
To obtain the normalized version of or , we divide by its Euclidean norm as:
We add a "hat" to the normalized vector to distinguish it from the un-normalized version.
Let's try an example with NumPy. I'll define vectors , compute its Euclidean norm, and then perform element-wise division :
x, y = np.array([[3],[4],[0]]), np.array([[-4],[3],[2]])
# euclidean norm of x and y
x_norm = np.linalg.norm(x, 2)
y_norm = np.linalg.norm(y, 2)
# normalized x or unit vector
x_unit = x * (1/x_norm)
y_unit = y * (1/y_norm)
print(f'Euclidean norm of x:\n{x_norm}\n')
print(f'Euclidean norm of y:\n{y_norm}\n')
print(f'Normalized x:\n{x_unit}\n')
print(f'Normalized y:\n{y_unit}')
"""
Euclidean norm of x:
5.0
Euclidean norm of y:
5.385164807134504
Normalized x:
[[0.6]
[0.8]
[0. ]]
Normalized y:
[[-0.74278135]
[ 0.55708601]
[ 0.37139068]]
"""
We can confirm that the Euclidean norm of the normalized versions of and equals by:
print(f'Euclidean norm of normalized x:\n{np.round(np.linalg.norm(x_unit, 2),1)}\n')
print(f'Euclidean norm of normalized y:\n{np.round(np.linalg.norm(y_unit, 2),1)}')
"""
Euclidean norm of normalized x:
1.0
Euclidean norm of normalized y:
1.0
"""
Taking and as a set, we can confirm the conditions for the definition of orthonormal vectors are correct.
print(f'Inner product normalized vectors:\n{np.round(x_unit.T @ y_unit,1)}\n')
print(f'Inner product normalized x with itself:\n{np.round(x_unit.T @ x_unit,1)}\n')
print(f'Inner product normalized y with itself:\n{np.round(y_unit.T @ y_unit,1)}')
"""
Inner product normalized vectors:
[[-0.]]
Inner product normalized x with itself:
[[1.]]
Inner product normalized y with itself:
[[1.]]
"""
Sets of vectors can be represented as matrices. We denote as the special case of a matrix composed of orthonormal vectors. The same properties we defined for sets of vectors hold when represented in matrix form.
Orthonormal basis transpose
A nice property of is that the matrix product with its transpose equals the identity:
This is true even when is not square. Let's see this with the orthonormal matrix resulting from stacking and .
Q = np.column_stack((x_unit, y_unit))
print(f'Orthonormal matrix Q:\n{Q}')
"""
Orthonormal matrix Q:
[[ 0.6 -0.74278135]
[ 0.8 0.55708601]
[ 0. 0.37139068]]
"""
Now we confirm
np.round(Q.T @ Q,1)
"""
array([[ 1., -0.],
[-0., 1.]])
"""
This property will be useful for several applications. For instance, the coupling matrix or correlation matrix of a matrix equals . If we are able to transform the vectors of into orthonormal vectors, such expressions reduces to . Other applications are the Fourier series and Least Square problems (as we will see later).
Gram-Schmidt Orthogonalization
In the previous section, I selected orthogonal vectors to illustrate the idea of an orthonormal basis. Unfortunately, in most cases, matrices are not full rank, i.e., not composed of a set of orthogonal vectors. Fortunately, there are ways to transform a set of non-orthogonal vectors into orthogonal vectors. This is the so-called Gram-Schmidt orthogonalization procedure.
The Gram-Schmidt orthogonalization consist of taking the vectors of a matrix, one by one, and making each subsequent vector orthonormal to the previous one. This is easier to grasp with an example. Consider the matrix :
What we want to do, is to find the set of orthonormal vectors , starting from the columns of , i.e., . We can select any vector to begin with. Recall that we normalize vectors by dividing by its norm as:
Let's approach this with NumPy:
A = np.array([[2, 1, -2],
[7, -3, 1],
[-3, 5, -1]])
A simple way to check the columns of are not orthonormal is to compute , which should be equal to the identity in the orthonormal case.
A.T @ A
"""
array([[ 62, -34, 6],
[-34, 35, -10],
[ 6, -10, 6]])
"""
To build our orthogonal set, we begin by denoting as .
Our first step is to generate the vector from such that is orthogonal to (i.e., ). To do this, we start with and subtract its projection along , which yields the following expression:
Think in this expression carefully. What are we doing, is to subtract times the first column from the second column. Let's denote as , then, we can rewrite our expression as:
As we will see, is a scalar, so effectively we are substracting an scaled version of column one from column two. The figure below express geometrically, what I have been saying: the non-orthogonal is projected onto . Then, we subtract the projection from to obtain which is orthogonal to as you can appreciate visually (recall ).
Keep these ideas in mind as it will be important later for QR decomposition.
Let's compute now:
q1 = A[:, 0]
a2 = A[:, 1]
q2 = a2 - ((q1.T @ a2)/(q1.T @ q1)) * q1
Let's check that and are actually orthogonal. If so, their dot product should be .
np.round(q1 @ q2, 2)
# -0.0
Next, we need to generate from . This time, we want to be orthogonal to both and . Therefore, we need to subtract its projection along and , which yields:
a3 = A[:, 2]
q3 = a3 - (((q1.T @ a3)/(q1.T @ q1)) * q1) - (((q2.T @ a3)/(q2.T @ q2)) * q2)
Verify orthogonality
print(f'Dot product q1 and q3:\n{np.round(q1 @ q3, 1)}\n')
print(f'Dot product q2 and q3:\n{np.round(q2 @ q3, 1)}')
"""
Dot product q1 and q3:
-0.0
Dot product q2 and q3:
0.0
"""
We can put into :
Q_prime = np.column_stack((q1, q2, q3))
print(f'Orthogonal matrix Q:\n{Q_prime}')
"""
Orthogonal matrix Q:
[[ 2. 2.09677419 -1.33333333]
[ 7. 0.83870968 0.66666667]
[-3. 3.35483871 0.66666667]]
"""
The reason we call this matrix is that although vectors are orthogonal, they are not normal.
Q_norms = np.linalg.norm(Q_prime, 2, axis=0)
print(f'Norms of vectors in Q-prime:\n{Q_norms}')
"""
Norms of vectors in Q-prime:
[7.87400787 4.04411161 1.63299316]
"""
We rename to by normalizing its vectors.
Q = Q_prime / Q_norms
np.linalg.norm(Q , 2, axis=0)
# array([1., 1., 1.])
To confirm we did this right, let's evaluate , that should return the identity:
np.round(Q.T @ Q, 1)
"""
array([[ 1., -0., -0.],
[-0., 1., 0.],
[-0., 0., 1.]])
"""
There you go: we performed Gram-Schmidt orthogonalization of
QR decomposition as Gram-Schmidt Orthogonalization
Gaussian Elimination can be represented as LU decomposition. Similarly, Gram-Schmidt Orthogonalization can be represented as QR decomposition.
We learned is an orthonormal matrix. Now let's examine , which is an upper triangular matrix. In LU decomposition, we used elementary matrices to perform row operations. Similarly, in the case of QR decomposition, we will use elementary matrices to perform column operations. We used a lower triangular matrix to perform row operations in LU decomposition by multiplying from the left side. This time, we will use an upper triangular matrix to perform column operations in QR decomposition by multiplying from the right side.
Once again, our starting point is the identity matrix. The idea is to alter the identity with the operations we want to perform over . Consider the matrix from our previous example:
What we did in out first step, wast to subtract of column from column . Let's compute first:
A = np.array([[2, 1, -2],
[7, -3, 1],
[-3, 5, -1]])
a1 = A[:, 0]
a2 = A[:, 1]
alpha = (a1.T @ a2)/(a1.T @ a1)
print(f'alpha factor:{np.round(alpha, 2)}')
# alpha factor:-0.55
Now we need to subtract times from . We can represent this operation with an elementary matrix, by doing applying the same operations the identity:
Next, we have to subtract times column and times from . Let's compute the new and :
a3 = A[:, 2]
beta = (a1.T @ a3)/(a1.T @ a1)
gamma = (a2.T @ a3)/(a2.T @ a2)
print(f'beta factor:{np.round(beta, 2)}')
print(f'gamma factor:{np.round(gamma, 2)}')
"""
beta factor:0.1
gamma factor:-0.29
"""
We can add this operations to our elementary matrix by subtracting times the first column from the third, and times the second from the third:
The last step is to normalize each vector of
l = np.array([[1, alpha, beta],
[0, 1, gamma],
[0, 0, 1]])
At this point, we should be able to recover :
As the matrix product of and
print(f'Q-prime and l product:\n{np.round(Q_prime @ l)}')
"""
Q-prime and l product:
[[ 2. 1. -2.]
[ 7. -3. 1.]
[-3. 5. -1.]]
"""
It works! Now, to recover will be difficult because of numerical stability and approximation issues in how we have computed things. Actually, if you remove the rounding from np.round(Q_prime @ l) you will obtain different numbers. Fortunately, there is no need to compute and by hand. We follow the previous steps merely for pedagogical purposes. In NumPy, we can compute the QR decomposition as:
Q_1, R = np.linalg.qr(A)
Let's compare our with
print(f'Q:\n{Q}\n')
print(f'Q:\n{Q_1}')
"""
Q:
[[ 0.25400025 0.51847585 -0.81649658]
[ 0.88900089 0.20739034 0.40824829]
[-0.38100038 0.82956136 0.40824829]]
Q:
[[-0.25400025 0.51847585 -0.81649658]
[-0.88900089 0.20739034 0.40824829]
[ 0.38100038 0.82956136 0.40824829]]
"""
The numbers are the same, but some signs are flipped. This stability and approximation issues is why you probably always want to use NumPy functions (when available).
Determinant
If matrices had personality, the determinant would be the personality trait that reveals most information about the matrix character. The determinant of a matrix is a single number that tells whether a matrix is invertible or singular, this is, whether its columns are linearly independent or not, which is one of the most important things you can learn about a matrix. Actually, the name "determinant" refers to the property of "determining" if the matrix is singular or not. Specifically, for an square matrix , a determinant equal to , denoted as , implies the matrix is singular (i.e., noninvertible), whereas a determinant equal to , denoted as , implies the matrix is not singular (i.e., invertible). Although determinants can reveal if matrices are singular with a single number, it's not used for large matrices as Gaussian Elimination is faster.
Recall that matrices can be thought of as function action on vectors or other matrices. Thus, the determinant can also be considered a linear mapping of a matrix onto a single number. But, what does that number mean? So far, we have defined determinants based on their utility of determining matrix invertibility. Before going into the calculation of determinants, let's examine determinants from a geometrical perspective to gain insight into the meaning of determinants.
Determinant as measures of volume
From a geometric perspective, determinants indicate the area of a parallelogram (e.g., a rectangular area) and the volume of the parallelepiped, for a matrix whose columns consist of the basis vectors in Euclidean space.
Let's parse out the above phrase: the area indicates the absolute value of the area, and the volume equals the absolute value of the volume. You may be wondering why we need to take the absolute value since real-life objects can't have negative area or volume. In linear algebra, we say the area of a parallelogram is negative when the vectors forming the figure are clockwise oriented (i.e., negatively oriented), and positive when the vectors forming the figure are counterclockwise oriented (i.e., positively oriented).
Here is an example of a matrix with vectors clockwise or negatively oriented:
The elements of the first column, indicate the first vector of the matrix, while the elements of the second column, the second vector of the matrix. Therefore, when we measure the area of the parallelogram formed by the pair of vectors, we move from left to right, i.e., clockwise, meaning that the vectors are negatively oriented, and the area of the matrix will be negative.
Here is the same matrix with vectors counterclockwise or positively oriented:
Again, the elements of the first column, indicate the first vector of the matrix, while the elements of the second column, the second vector of the matrix. Therefore, when we measure the area of the parallelogram formed by the pair of vectors, we move from right to left, i.e., counterclockwise, meaning that the vectors are positively oriented, and the area of the matrix will be positive.
The figure below exemplifies what I just said.
The situation for the volume of the parallelepiped is no different: when the vectors are counterclockwise oriented, we say the vectors are positively oriented (i.e., positive volume); when the vectors are clockwise oriented, we say the vectors are negatively oriented (i.e., negative volume).
The 2 X 2 determinant
Recall that matrices are invertible or nonsingular when their columns are linearly independent. By extension, the determinant allow us to whether the columns of a matrix a linearly independent. To understand this method, let's examine the special case first.
Consider a square matrix as:
How can we decide whether the columns are linearly independent? A strategy that I often use in simple cases like this, is just to examine whether the second column equals the first column times some factor. In the case of is easy to see that the second column equals four times the first column, so the columns are linearly dependent. We can express such criteria by comparing the elementwise division between each element of the second column by each element of the first column as:
We obtain that both entries equal , meaning that the second column can be divided exactly by the first column (i.e., linearly dependent).
Consider this matrix now:
Let's try again our method for :
Now we got into a problem because division by is undefined, so we can determine the relationship between columns of . Yet, by inspection, we can see the first column is simply times the second column, therefore linearly dependent. Here is when determinants come to the rescue.
Consider the generic matrix:
According to our previous strategy, we had:
This is, we tested the elementwise division of the second column by the first column. Before, we failed because of division, so we probably want a method that does not involve it. Notice that we can rearrange our expression as:
Let's try again with this method for :
And for :
It works. Indeed, are equal for both matrices, and , meaning their columns are linearly dependent. Finally, notice that we can rearange all the terms on one side of the equation as:
There you go: the above expression is what is known as the determinant of a matrix. We denote the determinant as:
Or
The N X N determinant
As matrices larger, computing the determinant gets more complicated. Consider the case as:
The problem now is that linearly independent columns can be either: (1) multiples of another column, and (2) linear combinations of pairs of columns. The determinant for a is:
Such expression is hard to memorize, and it will get even more complicated for larger matrices. For instance, the entails 24 terms. As with most things in mathematics, there is a general formula to express the determinant compactly, which is known as the Leibniz's formula:
Where computes the permutation for the rows and columns vectors of the matrix. Is of little importance for us to break down the meaning of this formula since we are interested in its applicability and conceptual value. What is important to notice, is that for an arbitrary square matrix, we will have terms to sum over. For instance, for a matrix, , which is a gigantic number considering the size of the matrix. In machine learning, we care about matrices with thousands or even millions of columns, so there is no use for such formula. Nonetheless, this does not mean that the determinant is useless, but the direct calculation with the above algebraic expression is not used.
Determinants as scaling factors
When we think in matrices as linear mappings, this is, as functions applied to vectors (or vectors spaces), the determinant acquires an intuitive geometrical interpretation: as the factor by which areas are scaled under a mapping. Plainly, if you do a mapping or transformation, and the area increases by a factor of , then the determinant of the transformation matrix equals . Consider the matrix and the basis vector :
Is easy to see that the parallelogram formed by the basis vectors of is . When we apply , we get:
A = np.array([[4, 0],
[0, 3]])
x = np.array([1,1])
A @ x.T
# array([4, 3])
Meaning that the vertical axis was scaled by and the horizontal axis by , hence, the new parallelogram has area . Since the new area has increased by a factor of , the determinant . Although we exemplified this with the basis vectors in , the determinant of for mappings of the entire vector space. The figure below visually illustrates this idea.
The importance of determinants
Considering that calculating the determinant is not computationally feasible for large matrices and that we can determine linear independence via Gaussian Elimination, you may be wondering what's the point of learning about determinants in the first place. I also asked myself more than once. It turns out that determinants play a crucial conceptual role in other topics in matrix decomposition, particularly eigenvalues and eigenvectors. Some books I reviewed devote a ton of space to determinants, whereas others (like Strang's Intro to Linear Algebra) do not. In any case, we study determinants mostly because of its conceptual value to better understand linear algebra and matrix decomposition.
Eigenthings
Eigenvectors, eigenvalues, and their associated mathematical objects and properties (which I call "Eigen-things") have important applications in machine learning like Principal Component Analysis (PCA), Spectral Clustering (K-means), Google's PageRank algorithm, Markov processes, and others. Next, we will review several of these "eigen-things".
Change of basis
Previously, we said that a set of linearly independent vectors with elements forms a basis for a vector space. For instance, we say that the orthogonal pair of vectors and (or horizontal and vertical axes), describe the Cartesian plane or space. Further, if we think in the and pair as unit vectors, then we can describe any vector in as a linear combination of and . For example, the vector:
Can be described as scaling the unit vector by , and scaling the unit vector by .
If you are like me, you have probably gotten use to the idea of describing any 2-dimensional space as and coordinates, with lying perfectly horizontal and perpendicular to it, as if this were the only natural way of thinking on coordinates in space. It turns out, there is nothing "natural" about it. You could literally draw a pair of orthogonal vectors on any orientation in space, define the first one as , and the second one as , and that would be perfectly fine. It may look different, but every single mathematical property we have studied so far about vectors would hold. For instance, in Fig 19. the alternative coordinates and are equivalent to the vectors and , in the standard and coordinates.
The question now is how to "move" from one set of basis vectors to the other. The answer is with linear mappings. We know already that equals to and in coordinates. To find the values of in , we need to take the inverse of . Think about it in this way: we represented in by scaling its unit vectors by the transformation matrix as:
Now, to do the opposite, i.e., to "translate" the values of the coordinates to values in , we scale by the inverse of as:
# Transformation or mapping matrix
T = np.array([[2, -2],
[2, 2]])
# Inverse of T
T_i = np.linalg.inv(T)
# x, y vectors
A = np.array([[1, 0],
[0,1]])
print(f"x, y vectors in x', y' coordinates:\n{T_i @ A}")
"""
x, y vectors in x', y' coordinates:
[[ 0.25 0.25]
[-0.25 0.25]]
"""
That is our answer: equals to and in coordinate space. Fig. 19 illustrate this as well.
This may become clearer by mapping in , onto in alternative coordinates. To do the mapping, again, we need to multiply by . Let's try this out with NumPy:
# vector to map
a = np.array([[-3],[-1]])
print(f'Vector a=[1,3] in x\' and y\' basis:\n{T_i@a}')
"""
Vector a=[1,3] in x' and y' basis:
[[-1. ]
[ 0.5]]
"""
In Fig. 19, we can confirm the mapping by simply visual inspection.
Eigenvectors, Eigenvalues and Eigenspaces
Eigen is a German word meaning "own" or "characteristic". Thus, roughly speaking, the eigenvector and eigenvalue of a matrix refer to their "characteristic vector" and "characteristic value" for that vector, respectively. As a cognitive scientist, I like to think in eigenvectors as the "pivotal" personality trait of someone, i.e., the personality "axis" around which everything else revolves, and the eigenvalue as the "intensity" of that trait.
Put simply, the eigenvector of a matrix is a non-zero vector that only gets scaled when multiplied by a transformation matrix . In other words, the vector does not rotate or change direction in any manner. It just gets larger or shorter. The eigenvalue of a matrix is the factor by which the eigenvector gets scaled. This is a bit of a stretch, but in terms of the personality analogy, we can think in the eigenvector as the personality trait that does not change even when an individual change of context: Lisa Simpson "pivotal" personality trait is conscientiousness, and no matter where she is, home, school, etc., their personality revolves around that. Following the analogy, the eigenvalue would represent the magnitude or intensity of such traits in Lisa. Fig 20. illustrate the geometrical representation of an eigenvector with a cube rotation.
More formally, we define eigenvectors and eigenvalues as:
Where is a square matrix in , the eigenvector, and an scalar in . This identity may look weird to you: How do we go from matrix-vector multiplication to scalar-vector multiplication? We are basically saying that somehow multiplying by a matrix or a scalar yields the same result. To make sense of this, recall our discussion about the effects of a matrix on a vector. Mappings or transformation like reflection and shear boils down to a combination of scaling and rotation. If a mapping does not rotate , it makes sense that such mapping can be reduced to a simpler scalar-vector multiplication .
If you recall our discussion about elementary matrices, you may see a simple way to make the more intuitive. Elementary matrices allow us to encode row and column operations on a matrix. Scalar multiplication, can be represented as by multiplying either the rows or columns of the identity matrix by the desired factor. For instance, for :
This allow us to rewrite as , and to maintain the matrix-vector multiplication form as:
We can go further, and rearange our expression to:
And to factor our to get:
The first part of our new expression, , will yield a matrix, meaning that now we have matrix-vector multiplication. In particular, we want a non-zero vector that when multiplied by yields . The only way to achieve this is when the scaling factor associated with is as well. Here is when determinants come into play. Recall that the determinant of a matrix represents the scaling factor of such mapping, which in this specific case, happens to be the eigenvalue of the matrix. Consequently, we want:
Since and are fixed, in practice, we want to find a value of that will yield a determinant of the matrix. Any matrix with a determinant of will be singular. This time, we want the matrix to be singular, as we are trying to solve a problem with three unknowns and two equations, therefore, it is the only way to solve it.
By finding a value for that makes the determinat , we are effectively making the equality true.
Let's do an example to make these ideas more concrete. Consider the following matrix:
Let's first multiply to get a single matrix:
We begin by computing the determinant as:
Which yield the following polynomial:
That we solve as any other quadratic polynomial, which receives the special name of characteristc polynomial. When we equate the characteristic polynomial to , we call such expression the characteristic equation. The roots of the characteristic equation, are the eigenvalues of the matrix:
Wich can be factorized as:
There you go: we obtain eigenvalues , and . this simply means that can be solved for eigenvalues equal to and , assuming non-zero eigenvectors.
Once we find the eigenvalues, we can compute the eigenvector for each of them. Let's start with :
Since the first and second column are identical, we obtain that the solution for the system is pair of such that , for instance:
Such vector correspond to the eigenspace for the eigenvalue . An eigenspace denotes all the vectors that correspond to a given eigenvalue, which in this case is the span of .
Now let's evaluate for :
Since the first column is just times the second, the solution for the system will be any pair such that , i.e.:
With the span of as the eigenspace for the eigenvalue .
As usual, we can find the eigenvectors and eigenvalues of a matrix with NumPy. Let's check our computation:
A = np.array([[4, 2],
[1, 3]])
values, vectors = np.linalg.eig(A)
print(f'Eigenvalues of A:\n{values}\n')
print(f'Eigenvectors of A:\n{np.round(vectors,3)}')
"""
Eigenvalues of A:
[5. 2.]
Eigenvectors of A:
[[ 0.894 -0.707]
[ 0.447 0.707]]
"""
The eigenvalues are effectively and . The eigenvectors (aside rounding error), match exactly what we found. For , , and for that .
Not all matrices will have eigenvalues and eigenvectors in . Recall that we said that eigenvalues essentially indicate scaling, whereas eigenvectors indicate the vectors that remain unchanged under a linear mapping. It follows that if a linear transformation does not stretch vectors and rotates all of them, then no eigenvectors and eigenvalues should be found. An example of this is a rotation matrix:
Let's compute its eigenvectors and eigenvalues in NumPy:
B = np.array([[0, -1],
[1, 0]])
values, vectors = np.linalg.eig(B)
print(f'B Eigenvalues:\n{values}\n')
print(f'B Eigenvectors:\n{vectors}\n')
"""
B Eigenvalues:
[0.+1.j 0.-1.j]
B Eigenvectors:
[[0.70710678+0.j 0.70710678-0.j ]
[0. -0.70710678j 0. +0.70710678j]]
"""
The +0.j indicates the solution yield imaginary numbers, meaning that there are not eigenvectors or eigenvalues for the matrix
Trace and determinant with eigenvalues
The trace of a matrix is the sum of its diagonal elements. Formally, we define the trace for a square matrix as:
There is something very special about eigenvalues: its sum equals the trace of the matrix. Recall the matrix from the previous section:
Which has a trace equal to . We found that their eigenvalues were and , which also add up to .
Here is another curious fact about eigenvalues: its product equals to the determinant of the matrix. The determinant of equals to . The product of the eigenvalues is also .
These two properties hold only when we have a full set of eigenvalues, this is when we have as many eigenvalues as dimensions in the matrix.
Eigendecomposition
In previous sections, we associated LU decomposition with Gaussian Elimination and QR decomposition with Gram-Schmidt Orthogonalization. Similarly, we can associate the Eigenvalue algorithm to find the eigenvalues and eigenvectors of a matrix, wit the Eigendecomposition or Eigenvalue Decomposition.
We learned that we can find the eigenvalues and eigenvectors of a square matrix (assuming they exist) with:
Process that entail to first solve the characteristic equation for the polynomial, and then evaluate each eigenvalue to find the corresponding eigenvector. The question now is how to express such process as a single matrix-matrix operation. Let's consider the following transformation matrix:
Let's begin by computing the eigenvalues and eigenvectors with NumPy:
A = np.array([[5, 3, 0],
[2, 6, 0],
[4, -2, 2]])
eigenvalues, eigenvectors = np.linalg.eig(A)
print(f'B Eigenvalues:\n{eigenvalues}\n')
print(f'B Eigenvectors:\n{eigenvectors}\n')
"""
B Eigenvalues:
[2. 8. 3.]
B Eigenvectors:
[[ 0. 0.6882472 0.18291323]
[ 0. 0.6882472 -0.12194215]
[ 1. 0.22941573 0.97553722]]
"""
We obtained a vector of eigenvalues and a Now, we know that the following identity must be true for scalar-matrix multiplication:
Since we want to multiply a matrix of eigenvalues by the matrix of eigenvectors, we have to be careful about selecting the order of the multiplication. Recall that matrix-matrix multiplication is not commutative, meaning that the multiplication order matters. Before this wasn't a problem, because scalar-matrix multiplication is commutative. What we want, is in operation such that eigenvalues scale eigenvectors. For this, we will put the eigenvectors in a matrix , the result of in a matrix , and multiply by from the right side as:
Let's do this with NumPy
X = eigenvectors
I = np.identity(3)
L = I * eigenvalues
print(f'Left-side of the equation AX:\n{A @ X}\n')
print(f'Right-side of the equation XL:\n{X @ L}\n')
"""
Left-side of the equation AX:
[[ 0. 5.50597761 0.54873968]
[ 0. 5.50597761 -0.36582646]
[ 2. 1.83532587 2.92661165]]
Right-side of the equation XL:
[[ 0. 5.50597761 0.54873968]
[ 0. 5.50597761 -0.36582646]
[ 2. 1.83532587 2.92661165]]
"""
Verify equality
print(f'Entry-wise comparison: {np.allclose(A @ X, X @ L)}')
# Entry-wise comparison: True
A a side note, it is not a good idea to compare NumPy arrays with the equality operator, as rounding error and the finite internal bit representation may yield False when values are technically equal. For instance:
(A @ X == X @ L)
"""
array([[ True, True, False],
[ True, True, False],
[ True, True, True]])
"""
We still have one issue to address to complete the Eigendecomposition of : to get rid of on the left side of the equation. A first thought is simply to multiply by the to cancel on both sides. This won't work because on the left side of the equation, is multiplying from the right of , whereas on the right side of the equation, is multiplying from the left of . Yet, we still can get take the inverse to eliminate only from the left side of the equation and obtain:
Lo and behold, we have found the expression for the Eigendecomposition.
Let's confirm this works:
X_inv = np.linalg.inv(X)
print(f'Original matrix A:\n{A}\n')
print(f'Reconstruction of A with Eigen Decomposition of A:\n{X @ L @ X_inv}')
"""
Original matrix A:
[[ 5 3 0]
[ 2 6 0]
[ 4 -2 2]]
Reconstruction of A with Eigen Decomposition of A:
[[ 5. 3. 0.]
[ 2. 6. 0.]
[ 4. -2. 2.]]
"""
Eigenbasis are a good basis
There are cases when a transformation or mapping has associated a full set of eigenvectors, i.e., as many eigenvectors as dimensions in . We call this set of eigenvectors an eigenbasis.
When approaching linear algebra problems, selecting a "good" basis for the matrix or vector space can significantly simplify computation, and also reveals several facts about the matrix that would be otherwise hard to see. Eigenbasis, are an example of a basis that would make our life easier in several situations.
From the previous section, we learned that the Eigenvalue Decomposition is defined as:
Conceptually, a first lesson is that transformations, like , have two main components: a matrix that stretch, shrink, or flip, the vectors, and , which represent the "axes" around which the transformation occurs.
Eigenbasis also make computing the power of a matrix easy. Consider the case of :
Since equals the identity, we obtain:
The pattern:
Generalizes to any power. For powers of or such approach may not be the best, as computing the power directly on may be easier. But, when dealing with large matrices with powers of thousands or millions, this approach is far superior. Further, it even works for the inverse:
We can see this is true by testing that equals the identity:
Pay attention to what happens now: , which yields:
Now, , also yields the identity:
Finally, , also yields the identity:
Geometric interpretation of Eigendecomposition
We said that Eigenbasis is a good basis as it allows us to perform computations more easily and to better understand the nature of linear mappings or transformations. The geometric interpretation of Eigendecomposition further reinforces that point. In concrete, the Eigendecomposition elements can be interpreted as follow:
- change basis (rotation) from the standard basis into the eigenbasis
- scale (stretch, shrinks, or flip) the corresponding eigenvectors
- change of basis (rotation) from the eigenbasis basis onto the original standard basis orientation
Fig 21. illustrate the action of , , and in a pair of vectors in the standard basis.
The problem with Eigendecomposition
The problem is simple: Eigendecomposition can only be performed on square matrices, and sometimes the decomposition does not even exist. This is very limiting from an applied perspective, as most practical problems involve non-square matrices.
Ideally, we would like to have a more general decomposition, that allows for non-square matrices and that exist for all matrices. In the next section we introduce the Singular Value Decomposition, which takes care of these issues.
Singular Value Decomposition
Singular Value Decomposition (SVD) is one the most relevant decomposition in applied settings, as it goes beyond the limitations of Eigendecomposition. Specifically, SVD can be performed for non-squared matrices and singular matrices (i.e., matrices without a full set of eigenvectors). SVD can be used for the same applications that Eigendecomposition (e.g., low-rank approximations) plus the cases for which Eigendecomposition does not work.
Singular Value Decomposition Theorem
Since we reviewed Eigendecomposition already, understanding SVD becomes easier. The SVD theorem states that any rectangular matrix can be decomposed as the product of an orthogonal matrix , a diagonal matrix , and another orthogonal matrix :
Another common notation is: . Here I'm using just to denote that the right orthogonal matrix is the same as in the Eigenvalue decomposition. Also notice that the inverse of an square orthogonal matrix is .
The Singular Values are the non-negative values along the diagonal of , which play the same role as eigenvalues in Eigendecomposition. You may even find some authors call them eigenvalues as well. Since is a rectangular matrix of the shape as , the diagonal of the matrix which contains the singular values will necessarily define a square submatrix within . There are two situations to pay attention to: (1) when , i.e., more rows than columns, and (2) when , i.e., more columns than rows.
For the first case, , we will have zero-padding at the bottom of as:
For the second case, , we will have zero-padding at the right of as:
Take the case of , the SVD is defined as:
Now let's evaluate the opposite case, , the SVD is defined as:
Singular Value Decomposition computation
SVD computation leads to messy calculations in most cases, so this time I'll just use NumPy. We will compute three cases: a wide matrix , a tall matrix , and a square matrix with a pair of linearly dependent vectors (i.e., a "defective" matrix, or singular, or not full rank, etc.).
# 2 x 3 matrix
A_wide = np.array([[2, 1, 0],
[-3, 0, 1]])
# 3 x 2 matrix
A_tall = np.array([[2, 1],
[-3, 0],
[0, 2]])
# 3 x 3 matrix: col 3 equals 2 x col 1
A_square = np.array([[2, 1, 4],
[-3, 0, -6],
[1, 2, 2]])
U1, S1, V_T1 = np.linalg.svd(A_wide)
U2, S2, V_T2 = np.linalg.svd(A_tall)
U3, S3, V_T3 = np.linalg.svd(A_square)
print(f'Left orthogonal matrix wide A:\n{np.round(U1, 2)}\n')
print(f'Singular values diagonal matrix wide A:\n{np.round(S1, 2)}\n')
print(f'Right orthogonal matrix wide A:\n{np.round(V_T1, 2)}')
"""
Left orthogonal matrix wide A:
[[-0.55 0.83]
[ 0.83 0.55]]
Singular values diagonal matrix wide A:
[3.74 1. ]
Right orthogonal matrix wide A:
[[-0.96 -0.15 0.22]
[-0. 0.83 0.55]
[ 0.27 -0.53 0.8 ]]
"""
As expected, we obtain a orthogonal matrix on the left, and a orthogonal matrix on the right. NumPy only returns the singular values along the diagonal instead of the matrix, yet it makes no difference regarding the values of the SVD.
print(f'Left orthogonal matrix for tall A:\n{np.round(U2, 2)}\n')
print(f'Singular values diagonal matrix for tall A:\n{np.round(S2, 2)}\n')
print(f'Right orthogonal matrix for tall A:\n{np.round(V_T2, 2)}')
"""
Left orthogonal matrix for tall A:
[[-0.59 -0.24 -0.77]
[ 0.8 -0.32 -0.51]
[-0.13 -0.91 0.38]]
Singular values diagonal matrix for tall A:
[3.67 2.13]
Right orthogonal matrix for tall A:
[[-0.97 -0.23]
[ 0.23 -0.97]]
"""
As expected, we obtain a orthogonal matrix on the left and a orthogonal matrix on the right. Notice that NumPy returns the singular values in descending order of magnitude. This is a convention you'll find in the literature frequently.
print(f'Left orthogonal matrix for square A:\n{np.round(U3, 2)}\n')
print(f'Singular values diagonal matrix for square A:\n{np.round(S3, 2)}\n')
print(f'Right orthogonal matrix for square A:\n{np.round(V_T3, 2)}')
"""
Left orthogonal matrix for square A:
[[-0.54 -0.2 -0.82]
[ 0.79 -0.46 -0.41]
[-0.29 -0.86 0.41]]
Singular values diagonal matrix for square A:
[8.44 1.95 0. ]
Right orthogonal matrix for square A:
[[-0.44 -0.13 -0.89]
[ 0.06 -0.99 0.12]
[ 0.89 0. -0.45]]
"""
Although column three is just two times column one (i.e., linearly dependent), we obtain the SVD for . Notice that the third singular value equals , which is a reflection of the fact that the third column just contains redundant information.
Geometric interpretation of the Singular Value Decomposition
As with Eigendecomposition, SVD has a nice geometric interpretation as a sequence of linear mappings or transformations. Concretely:
- change basis (rotation) from the standard basis into a set of orthogonal basis
- scale (stretch, shrinks, or flip) the corresponding orthogonal basis
- change of basis (rotation) from the new orthogonal basis onto some other orientation, i.e., not necessarily where we started.
The key difference with Eigendecomposition is in : instead of going back to the standard basis, performs a change of basis onto another direction.
Fig 22. illustrate the effect of , i.e., , , and , in a pair of vectors in the standard basis. The fact that the right orthogonal matrix has column vectors generates the third dimension which is orthogonal to the ellipse surface.
Singular Value Decomposition vs Eigendecomposition
The SVD and Eigendecomposition are very similar, so it's easy to get confused about how they differ. Here is a list of the most important ways on which both are different:
- The SVD decomposition exist for any rectangular matrix , while the Eigendecomposition exist only for square matrices .
- The SVD decomposition exists even if the matrix is defective, singular, or not full rank, whereas the Eigendecomposition does not have a solution in in such a case.
- Eigenvectors are orthogonal only for symmetric matrices, whereas the vectors in the and are orthonormal. Hence, represents a rotation only for symmetric matrices, whereas and are always rotations.
- In the Eigendecomposition, and are the inverse fo each other, whereas and in the SVD are not.
- The singular values in are always real and positive, which is not necessarily the case for in the Eigendecomposition.
- The SVD change basis in both the domain and codomain. The Eigendecomposition change basis in the same vector space.
- For symmetric matrices, , the SVD and Eigendecomposition yield the same results.
Matrix Approximation
In machine learning applications, it is common to find matrices with thousands, hundreds of thousands, and even millions of rows and columns. Although the Eigendecomposition and Singular Value Decomposition make matrix factorization efficient to compute, such large matrices can consume an enormous amount of time and computational resources. One common way to "get around" these issues is to utilize low-rank approximations of the original matrices. By low-rank we mean utilizing a subset of orthogonal vectors instead of the full set of orthogonal vectors, such that we can obtain a "reasonably" good approximation of the original matrix.
There are many well-known and widely use low-approximation procedures in machine learning, like Principal Component Analysis, Factor Analysis, and Latent Semantic analysis, and dimensionality reduction techniques more generally. Low-rank approximations are possible because in most instances, a small subset of vectors contains most of the information in the matrix, which is a way to say the most data points can be computed as linear combinations of a subset of orthogonal vectors.
Best rank-k approximation with SVD
So far we have represented the SVD as the product of three matrices, , , and . We can represent this same computation as a the sum of the matching columns of each of these components as:
Notice that each iteration of will generate a matrix , which then can be multiplied by . In other words, the above expression also equals:
In matrix notation, we can express the same idea as:
Now, we can approximate by taking the sum over values instead of values. For instance, for a square matrix with orthogonal vectors, we can compute an approximation with the orthogonal vectors as:
In practice, this means that we take orthogonal vectors from and , times singular values, which requires considerably less computation and memory than the matrix. We call this the best low-rank approximation simply because it takes the largest singular values, which account for most of the information. Nonetheless, we still a precise way to estimate how good is our estimation, for which we need to compute the norm for and , and how they differ.
Best low-rank approximation as a minimization problem
In the previous section, we mentioned we need to compute some norm for and , and then compare. This can be conceptualized as a error minimization problem, where we search for the smallest distance between and the low-rank approximation . For instance, we can use the Frobenius and compute the distance between and as:
Alternatively, we can compute the explained variance for the decomposition, where the highest the variance the better the approximation, ranging from to . We can perform the SVD approximation with NumPy and sklearn as:
from sklearn.decomposition import TruncatedSVD
A = np.random.rand(100,100)
SVD1 = TruncatedSVD(n_components=1, n_iter=7, random_state=1)
SVD5 = TruncatedSVD(n_components=5, n_iter=7, random_state=1)
SVD10 = TruncatedSVD(n_components=10, n_iter=7, random_state=1)
SVD1.fit(A)
SVD5.fit(A)
SVD10.fit(A)
"""
TruncatedSVD(algorithm='randomized', n_components=10, n_iter=7, random_state=1,
tol=0.0)
"""
print('Explained variance by component:\n')
print(f'SVD approximation with 1 component:\n{np.round(SVD1.explained_variance_ratio_, 2)}\n')
print(f'SVD approximation with 5 components:\n{np.round(SVD5.explained_variance_ratio_, 2)}\n')
print(f'SVD approximation with 10 component:\n{np.round(SVD10.explained_variance_ratio_,2)}')
"""
Explained variance by component:
SVD approximation with 1 component:
[0.01]
SVD approximation with 5 components:
[0.01 0.04 0.03 0.03 0.03]
SVD approximation with 10 component:
[0.01 0.04 0.03 0.03 0.03 0.03 0.03 0.03 0.03 0.03]
"""
print('Singular values for each approximation:\n')
print(f'SVD approximation with 1 component:\n{np.round(SVD1.singular_values_, 2)}\n')
print(f'SVD approximation with 5 components:\n{np.round(SVD5.singular_values_, 2)}\n')
print(f'SVD approximation with 10 component:\n{np.round(SVD10.singular_values_,2)}')
"""
Singular values for each approximation:
SVD approximation with 1 component:
[50.42]
SVD approximation with 5 components:
[50.42 5.47 5.26 5.16 5.08]
SVD approximation with 10 component:
[50.42 5.47 5.26 5.16 5.08 5.06 4.99 4.88 4.72 4.63]
"""
print('Total explained variance by each approximation:\n')
print('Singular values for each approximation:\n')
print(f'SVD approximation with 1 component:\n{np.round(SVD1.explained_variance_ratio_.sum(), 2)}\n')
print(f'SVD approximation with 5 components:\n{np.round(SVD5.explained_variance_ratio_.sum(), 2)}\n')
print(f'SVD approximation with 10 component:\n{np.round(SVD10.explained_variance_ratio_.sum(),2)}')
"""
Total explained variance by each approximation:
Singular values for each approximation:
SVD approximation with 1 component:
0.01
SVD approximation with 5 components:
0.15
SVD approximation with 10 component:
0.29
"""
As expected, the more components (i.e., the highest the rank of the approximation), the highest the explained variance.
We can compute and compare the norms by first capturing each matrix of the SVD as recovering , then compute the Frobenius norm of the difference between and .
from sklearn.utils.extmath import randomized_svd
U, S, V_T = randomized_svd(A, n_components=5, n_iter=10, random_state=5)
A5 = (U * S) @ V_T
print(f"Norm of the difference between A and rank 5 approximation:\n{np.round(np.linalg.norm(A - A5, 'fro'), 2)}")
"""
Norm of the difference between A and rank 5 approximation:
26.32
"""
This number is not very informative in itself, so we usually utilize the explained variance as an indication of how good is the low-rank approximation.
Some notes
Notation
- - these are vectors and x is less than y elementwise
- - matrices, is PSD
Linearity
- inner product
- like inner product if we collapsed into big vector
- linear
- symmetric
- gives angle back
- linear
- superposition
- proportionality
- bilinear just means a function is linear in 2 variables
- vector space
- closed under addition
- contains identity
- det - sum of products including one element from each row / column with correct sign
- absolute value = area of parallelogram made by rows (or cols)
- lin independent:
- cauchy-schwartz inequality:
- implies triangle inequality:
Matrix properties
- nonsingular = invertible = nonzero determinant = null space of zero
- only square matrices
- rank of mxn matrix- max number of linearly independent columns / rows
- rank==m==n, then nonsingular
- ill-conditioned matrix - matrix is close to being singular - very small determinant
- inverse
- orthogonal matrix: all columns are orthonormal
- preserves the Euclidean norm
- if diagonal, inverse is invert all elements
- inverting 3x3 - transpose, find all mini dets, multiply by signs, divide by det
- psuedo-inverse = Moore-Penrose inverse
- if A is nonsingular,
- if rank(A) = m, then must invert using
- if rank(A) = n, then must use
- inversion of matrix is
- inverse of psd symmetric matrix is also psd and symmetric
- if A, B invertible
- orthogonal matrix: all columns are orthonormal
- orthogonal complement - set of orthogonal vectors
- define R(A) to be range space of A (column space) and N(A) to be null space of A
- R(A) and N(A) are orthogonal complements
- dim = r
- dim = n-r
- dim = r
- dim = m-r
- adjoint - compute with mini-dets
- Schur complement of
Matrix calc
- overview: imagine derivative
- function f:
- gradient vector - partial derivatives with respect to each element of A (vector or matrix)
- gradient =
- these next 2 assume numerator layout (numerator-major order, so numerator constant along rows)
- function f:
- Jacobian matrix: - this is dim(f) x dim(x)
- function f:
- 2nd derivative is Hessian matrix
- 2nd derivative is Hessian matrix
- examples
- (if A symmetric, else )
- (if A symmetric, else )
- we can calculate derivs of quadratic forms by calculating derivs of traces
- useful result:
Norms
- def
- nonnegative
- definite f(x) = 0 iff x = 0
- proportionality (also called homogenous)
- triangle inequality
- properties
- convex
Vector norms
- norms:
- norm - number of nonzero elements (this is not actually a norm!)
- - Euclidean norm
- - also called Cheybyshev norm
- quadratic norms
- P-quadratic norm: where
- dual norm
- given a norm , dual norm
- dual of the dual is the original
- dual of Euclidean is just Euclidean
- dual of is
- dual of spectral norm is some of the singular values
Matrix norms
schatten p-norms: - note this is nice for organization but this p is never really mentioned
- p=1: nuclear norm = trace norm:
- p=2: frobenius norm = euclidean norm:
- like vector norm
- p=: spectral norm = -norm (of a matrix) =
entrywise norms
- sum-absolute-value norm (like vector )
- maximum-absolute-value norm (like vector )
operator norm
- let and be vector norms
- operator norm
- represents the maximum stretching that X does to a vector u
- if using p-norms, can get Frobenius and some others
Eigenstuff
Eigenvalues intro - strang 5.1
- nice viz
- elimination changes eigenvalues
- eigenvector application to diff eqs
- soln is exponential:
- eigenvalue eqn:
- yields characteristic polynomial
- eigenvalue properties
- 0 eigenvalue A is singular
- eigenvalues are on the main diagonal when the matrix is triangular
- expressions when
- defective matrices - lack a full set of eigenvalues
- positive semi-definite:
- basically these are always symmetric
- all eigenvalues are nonnegative
- if then A is positive semi definite (PSD)
- like it curves up
- Note:
- if then A is positive definite (PD)
- PD full rank, invertible
- PSD + symmetric can be written as Gram matrix
- if X full rank, then is PD
- PSD notation
- - set of symmetric matrices
- - set of PSD matrices
- - set of PD matrices
Strang 5.2 - diagonalization
- diagonalization = eigenvalue decomposition = spectral decomposition
- assume A (nxn) is symmetric
- Q := eigenvectors as columns, Q is orthonormal
- only diagonalizable if n independent eigenvectors
- not related to invertibility
- eigenvectors corresponding to different eigenvalues are lin. independent
- other Q matrices won't produce diagonal
- there are always n complex eigenvalues
- orthogonal matrix
- examples
- if X, Y symmetric,
- lets us easily calculate ,
- eigenvalues of are squared, eigenvectors remain same
- eigenvalues of are inverse eigenvalues
- eigenvalue of rotation matrix is
- eigenvalues for only multiply when A and B share eigenvectors
- diagonalizable matrices share the same eigenvector matrix S iff
- generalized eigenvalue decomposition - for 2 symmetric matrices
- ,
Strang 6.3 - singular value decomposition
- SVD for any nxp matrix:
- U columns (nxn) are eigenvectors of
- columns of V (pxp) are eigenvectors of
- r singular values on diagonal of (nxp) - square roots of nonzero eigenvalues of both and
- like rotating, scaling, and rotating back
- SVD ex.
- properties
- for PD matrices, ,
- for other symmetric matrices, any negative eigenvalues in become positive in
- applications
- very numerically stable because U and V are orthogonal matrices
- condition number of invertible nxn matrix =
- we can throw away columns corresponding to small
- pseudoinverse
Strang 5.3 - difference eqs and power
- compound interest
- solving for fibonacci numbers
- Markov matrices
- steady-state Ax = x
- corresponds to
- stability of
- stable if all eigenvalues satisfy
- neutrally stable if some
- unstable if at least one
- Leontief's input-output matrix
- Perron-Frobenius thm - if A is a positive matrix (positive values), so is its largest eigenvalue and every component of the corresponding eigenvector is also positive
- useful for ranking, etc.
- power method: want to find eigenvector corresponding to largest eigenvalue
- where is nonnegative
Epilogue
Linear algebra is an enormous and fascinating subject. These notes are just an introduction to the subject with machine learning in mind. I am no mathematician, and I have no formal mathematical training, yet, I greatly enjoyed writing this document. I have learned quite a lot by doing it and I hope it may help others that, like me, embark on the journey of acquiring a new skill by themselves, even when such effort may seem crazy to others.